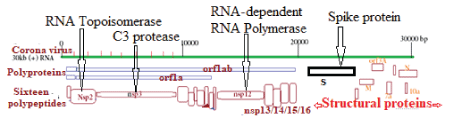
Figure 1A: Structure of human corona viruses (SARS CoV-2 or COVID-19).

Asit Kumar Chakraborty*
Department of Biotechnology and Biochemistry, Oriental Institute of Science and Technology, Midnapore, West Bengal, India*Corresponding author: Asit Kumar Chakraborty, Retired, Department of Biotechnology and Biochemistry, Oriental Institute of Science and Technology, Midnapore, West Bengal, India, Tel: +917679154141; E-mail: chakraakc@gmail.com
NCBI SARS-CoV-2 Database was analyzed between November-December, 2021 to decipher the spread of Delta corona virus variants in the USA and compared with highly transmissible new omicron variant recently originated in South Africa. Presently, B.1.617.2 and AY.103 lineages Delta variants with spike protein L452R, T478K, P681R mutations and F157/R158 two amino acids deletions were predominant in the USA and superseded the deadly outbreaks of B.1.1.7 Alpha variant with deletions of H69, V70 and Y145 amino acids as well as N501Y, and D614G highly transmissible mutations. Interestingly, omicron variant has six H69, V70, V143, Y144, Y145, L212 immune-escape deletions as well as 29 mutations in the spike protein including most deadly N501Y (Y498 in omicron) and D614G (G611 in omicron). This indicated that omicron variant was originated by combination among B.1.1.7, AY.X and B.1.617.2 lineages. A unique three amino acids (EPE) insertion at 215 position of spike protein was detected to compensate six deletions suggesting further recombination events. Three Serine residues were mutated at amino acids 371 (S=L, L368 in omicron), 373 (S=P, P370 in Omicron), 375 (S=F, F372 in omicron) but compensated at 446 (G=S, S443 in omicron) and 496 (G=S, S493 in omicron) at the RBD domain of omicron virus. The three amino acids (ERS) deletion at position 30 in the N-protein acts as another signature of omicron virus. Omicron variant has less mutation in the 2/3 5’-end of the genome that codes for ORF1ab poly-protein but dominant P4715L mutation in the RNA-dependent RNA polymerase. However, overall amino acid composition, alipathic index, and instability index were found fairly constant although hydrophobic plot gave some difference between spike protein of Wuhan and omicron corona viruses. BLAST search detected 20nt and 19nt perfect match of hyper-variable 22957-22977nt region comprising 488-493 amino acids (NH2-PLRSYS-CO2H) of the spike protein of omicron virus with the ch-2 of Seladonia tumulorum or ch-16 of Steromphala cineraria respectively. A primer set designed from the RBD domain of spike gene did not detected the omicron genome by BLAST search but primers from the constant regions of the genome worked well. Such hyper-variation in the spike protein suggested that DNA vaccine or mRNA vaccine using spike gene of corona virus may not efficiently protect omicron virus infection and attenuated whole corona virus vaccine will be safer vaccine.
NCBI SARS CoV-2 Database; Signatures of omicron corona virus; Hyper-variable spike protein; RT-PCR diagnosis; Vaccine failure; Antibody resistance
Since the December 2019 Wuhan corona virus (severe acute respiratory syndrome virus or SARS-CoV-2) has caused 0.58 million deaths worldwide with thousand mutations creating many dominant forms like alpha, delta, delta-plus and very recently omicron. Corona virus is a large positive-sense RNA virus with a compact 29,980 nucleotides-long genome and COVID-19 is related to six different corona viruses like CoV-229E, CoV-HKU1, CoV-OC43, CoV-NL63, SARS-CoV and Middle East respiratory syndrome corona virus (MERS-CoV) [1-5]. It has structural proteins (S, M, N, E) at the 3’- end and 5’ two very large poly-proteins (2/3 of the genome) which degraded into sixteen non-structural proteins (nsp1-16) including RNA-dependent RNA polymerase (nsp12) [6], two proteases (nsp3 and nsp5) [7,8], RNA topoisomerase (nsp2) [9], RNA helicase (nsp13) [10], nucleases (nsp15) [11] and methyl transferases (nsp16) [12] (figure 1A). Spike protein (1273 aa) is a trimeric class 1 transmembrane glycoprotein and its RBD domain (335-515 aa) acts as receptor binding domain to bind ACE-2 receptor of host cells for virus entry [13] (figure 1B). S protein 1-13 AA acts as signal peptide and the S1 subunit is 14-685 AA containing RBD domain and S2 subunit is 686 to 1273 amino acids with fusion contact peptide (788-806 AA) as well as two hepta-peptide (HPPHCPC) repeats at 1163 and 1213 positions [14]. Among the other structural proteins N-protein (419 aa) binds to leader RNA of replicating corona virus and also regulates host-pathogen interactions.

Figure 1A: Structure of human corona viruses (SARS CoV-2 or COVID-19).
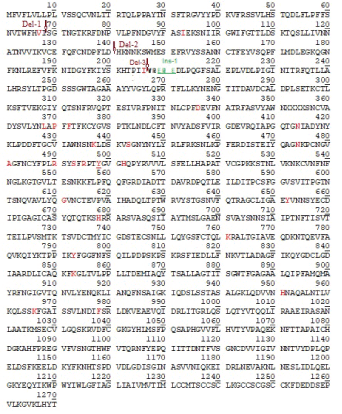
Figure 1B: Primary amino acid sequence of the spike protein of Omicron corona virus. Two deletion points were shown by Del-1 and Del-2 arrows, Insertion point was denoted by Ins-1 and stars. Red denotes mutant amino acids and with underlined means well characterized mutations that enhanced viral transmission (G611 and Y498 here).
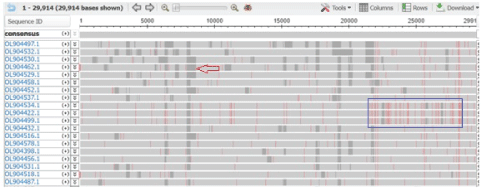
Figure 1C: Multi-alignment of 200-500 sequences at the NCBI SARS CoV-2 database showing gray gap (arrow) for incomplete sequence and heavy red lines for mutation in the spike protein between 21000-25000nt (blue box) that acted as signatures of omicron corona virus. Such signature is important to search omicron in the database where 200-500 sequences were aligned. Otherwise it was very hard to find omicron sequences. Interestingly, deposited many omicron sequences were incomplete in the RBD domain (313-393 aa/ 303-393 aa/ 425-439 aa) of the spike protein due to mutations.
Severe COVID-19 is more common in adults aged ~70 years with co-morbidities such as diabetes, cardiovascular disease and chronic respiratory disease. A difference in case fatality rates across countries was observed, possibly due to a diverse demographic composition and the type of control measures that have been taken in different countries to stop viral spreading [15]. According to 2020 database, three major Clades of SARS-CoV-2 can be identified and named as Clade G (variant of the spike protein S-D614G), Clade V (variant of the ORF3a coding protein NS3-G251V), Clade GR (S-D614G + N-G204R) and Clade S (variant ORF8-L84S) [16,17]. SARS-CoV-2 variants emerged many fold in late 2020, and at least three variants of concern (B.1.1.7, B.1.351, and P1) have been reported by WHO. Alpha variant B1.1.1.7 had caused havoc calamity in India, UK and USA between August 2020 and March 2021. Delta and Deltaplus variants specifically AY.103 and B.1.617.2 caused much fatality between March-December 2021. Delta spike triggers faster fusion with ACE-2 receptor of host lung cells relative to only D614G mutants suggesting greater pathogenicity of delta variants than B.1.1.7 lineage. Beta B.1.351, Gamma P.1, Epsilon B.1.427, Iota B.1.526, Mu B.1.621 and Zeta P.2 variants corona viruses were also indicated some concern in different demography [18]. However, recently omicron variant is highly spreading in South Africa and already has detected in 90 countries including Europe, USA, Australia and India. It has hyper-variable thirty spike mutations with important deletions and insertions. Thus, more complete sequences needed to define specific geographic distributions of omicron virus variant. Most importantly, clinical and political strategies at the local level must be augmented because spike gene DNA and mRNA vaccines may not work well for hyper-variable spike gene of omicron corona virus.
Presently, at least ten vaccine candidates vaccinated 70% world population. Vaccine usually is a protein or synthetic peptides from Coronavirus that can elicits humoral antibody (IgG) as well as T-cell mediated ability to destroy virus. Attenuated or killed Corona virus (Covaxin, Bharat Biotech, India) also used like Pox vaccination. As genetic information in cells processed from DNA to RNA to protein, scientists have exploited DNA vaccine as well as RNA vaccine for the protection of Corona virus. Indian Serum Institute uses killed virus where as Russia uses mRNA vaccine (Sputnik V) and England (Oxford + Astra-Zeneca) uses S gene DNA vaccine using adenovirus vector (Ad5 or Ad26). USA (Moderna/ Pfizer) and Germany’s BioNTech uses S gene mRNA vaccine [19]. The most companies used spike protein (S gene) which was the receptor protein of corona virus that bound to ACE-2 receptor of lung cells of human and animal.
Mutation greatly affected increase modes of virus transmission as in case of D614G and N501Y mutations [16]. Further, a decrease vaccine utility (protection against virus) was reported with immune escape (T cell immunity) as in case of 69, 70 and 145 amino acids deletions in alpha corona virus (B.1.1.7) [20]. Further, mutations like L452R, E484K, and other at the RBD of virus greatly lower the neutralization efficacy of serum antibody from earlier corona patients to mutant viruses [21]. Presently, deletion of F157 and R158 in AY.X and B.1.1.617.2 Delta variants produced increased transmission in presence of D614G mutation together further lowering the vaccine efficacy. Very recently, distinct omicron virus new mutations found lowering vaccine utility and increasing transmission rate but reports of confirmed immunological data yet to come [22-24]. We will molecularly study the spread of omicron virus specifically in the USA by analyzing NCBI Virus Database between 20th November to 25th December, 2021 using different free software available in the net.
We used NCBI (www.ncbi.nlm.gov) SARS-CoV-2 database only as it gave multi-alignment data for up to 500 sequences. Such alignment detected most sequences were incomplete and separately analyzed. But middle complete sequences were checked for comparable sequences by looking red lines for mutations and mostly AY.x variant corona viruses. Omicron viruses have many red lines between 21000-28000nt for the hyper-variable spike protein and other structural proteins (Figure 1C). Thus, we covered many sequences to few omicron virus sequences helpful for analysis by freely available Multalin software and CLUSTAL-Omega software. It took 2-3 minutes for spike protein (1273 aa) alignment by Multalin software but it took 30-40 minutes for CLUSTAL Omega software 30kb RNA genome alignment. As we found most sequences were AY.X Delta type (~85%) and some Delta B.1.1.617.2 type (~10%). We only documented data for omicron sequences (0.5-5%) deposited between 23rd November and 24th December, 2021. Date of sequence deposition in the NCBI Database, Author’s name and Collection date of virus were used to analyze sequence sets. Although first we detected one omicron corona virus sequence in such search. Then, we BLAST analyzed the hyper-variable regions (60 nt) to get more three omicron sequences but one US such US originated sequence had incomplete spike protein and two complete sequences each from Canada and Belgium. However, from December 6, more and more omicron viruses were detected in the database. Our guess sequencing primers used for Wuhan, Alpha and Delta variants were not worked well using standard kits available. But more and more omicron virus complete sequences will be deposited afterwards. We have no access to GISAIP database and the accession number for the first omicron virus genome (29684 bp) is EPI_ISL_6640916 with collection date 11-11-2021 and submission date 23-11-2021. Parts of multi-alignments were presented with different omicron signatures. During review of the paper, we also analyzed the database and huge omicron sequences were deposited in last week of December, 2021 and first week of January, 2022.
We first detected an omicron strain (B.1.1.529 or BA.1) of corona virus on 7th December by analyzing the NCBI virus database deposited on 6th December 2021 by Puehringer, et al., from Austria (accession no. OL721912; Date of isolation 1st December, 2021). We had not detected any omicron variants in US from data deposited by Bankers L, et al., and mostly found delta B.1.617.2 and AY.X strains (Figure 2). The 7th December 2021 deposited sequences by Howard D, et al., Schmedes, et al., Bankers L, et al., Pachucki R, et al., Buck GA, et al., Pokharel A, et al., Blankenship HM, et al. and Lighthouse Lab, et al., did not found any omicron as well. Then, we planned to BLAST search of 60nt (22894 5’- AAC TGA AAT CTA TCA GGC CGG TAA CAA ACC TTG TAA TGG TGT TGC AGG TTT TAA TTG TTA-3’ 22953) hyper-variable region of omicron corona virus genome. The study resulted three hyper-variable spike protein of omicron B.1.1.529 corona virus with accession nos. OL698718 (USA, incomplete S), OL677199 (Canada) and OL672836 (Belgium). We presented multialignment data to show mutations and deletions as compared with most deadly B.1.1.7, AY.103 and B.1.1617.2 variants (Figures 3A-3C). It proved that omicron variants had six H69, V70, E144, F145, R146 and L212 immune-escape deletions as well as 29 mutations in the spike protein including most deadly D614G (G611 in omicron virus) and N501Y (Y498 in omicron virus). Other mutations were: A67V (V67), T95I (I93), N211I (I206), L212V (V207), V215P (P210), R216E (E211), G341D (D336), S373L (L368), S375P (P370), S377F (F372), K419N (N414), N442K (K437), G448S (S443), S479N (N474), E486A (A481), Q495R (R490), G498S (S493), Q500R (R495), Y507H (H502), T549K (K544), H657Y (Y652), P683H (H678), N766K (K761), D798Y (Y793), N858K (K853), Q956H (H951), N971K (K966), and L983F (F978) [in sate values for omicron virus positions] (figure 1B and figures 3A-3C). The spike protein in Wuhan virus had 1273 aa, in B.1.1.7 variant had 1270 aa, in B.1.1.617.2 and AY.X variants had 1271 aa and in omicron variant had 1270 aa. The roles of those mutations were not clear yet and more research needed! P681R mutation (P683H disclosed here) was found in B.1.1.617.2 variant but no L452R mutation in omicron virus. It appeared EPE sequences inserted at 215 position and then substitutions would be happened. Receptor Binding Domain (RBD) of Spike protein bound to its ACE-2 receptor of human lung cells needed for virus entry. Analysis of ORF1ab protein (7096 aa long) suggested that dominant P4715L mutation in the RNA-dependent RNA polymerase and further K564N, K856R, L2084I, A2710T and P3395H mutations were happened in ORF1ab protein of omicron virus. P4715L, as well as L2084I mutations were also detected in Delta variants. However, S2083 deletion in omicron could be critical but no data available. In Delta variants other important mutations like A1306S, P2046L, P2287S, T2836I, V2930L, G5063S, P5401, A6319V, and K6958R were noticed (see, accession no. OL721909) as compared to Wuhan virus of December, 2019 (data not shown).
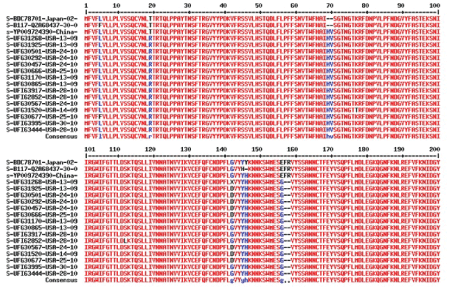
Figure 2: Most variant of corona virus detected in the USA before November 30, 2021 was AY.X Delta variant that superseded UK alpha B.1.1.7 variant.
We identified few spike protein conserved regions rich in hydrophobic amino acids (V, L, I, F) as indicated by green underline in figure 3B and figure 3C (green underlined) to explain a view that potentiality of Wuhan S gene vaccine for omicron virus neutralization as reported recently but much reduced efficacy (8x) would be possible. Thus, attenuated whole virus vaccine like Covaxin (India made) may be more important to control omicron corona virus spread until new S gene vaccine with omicron genome will be made.

Figure 3A: Deletions and insertions in the spike protein of omicron corona virus as compared with alpha and delta variants. Protein IDs UFP04971, UFO69279 and UFT26501 were omicron variant originated in Canada, Belgium and Austria respectively. Protein ID UFT26468 was B.1.617.2 Delta variant and highly active worldwide.

Figure 3B: Hyper-variable regions of RBD domain of spike protein of Omicron variant corona virus as compared with Alpha and Delta variants. Three Serine residues at 373, 375, and 377 were changed in omicron variants and compensated by G448S and G498S mutations. Conserved hydrophobic region was shown by green underline.
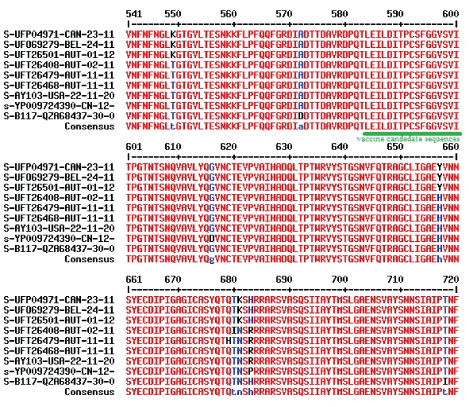
Figure 3C: Universal D614G mutation (100% now) of deadly corona viruses and new H657Y, N681K and P683H mutations in omicron variant. Conserved hydrophobic region was shown by green underline.
We continued our analysis as more new omicron sequences were deposited. Data deposited on 10-12-2021 by Lemieux JE, et al., and Howard D, et al., did not produce any omicron but mostly AY.X and some B.1.1.617.2 variants. However, data deposited on 11-12-2021 by Holland SL, et al., Howard D, et al., and Pinet K, et al., produced fourteen omicron variants between November 24 to December 6, 2021 collection dates. The accession numbers were: OL815080/81 (USA, AZ), OL815417 (USA, TX), OL815350/51 (USA, MA), OL815447/48 (USA, GA), OL815449/50 (USA, NY), OL8154/51/52/53 (USA, CA), and OL815455/56 (USA, MA). Multi-alignment produced very similar result as depicted in figure 4. Panel C data was not adjusted where RE insertion appeared not EPE due to unability of Multalin software to detect deletion L212. We kept that data because CLUSTALOmega protein multi-alignment also did not correct the data which we confirmed by DNA alignment as shown in figure 5.
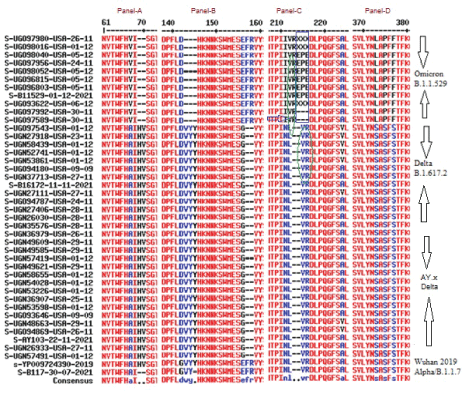
Figure 4: Database Analysis of dated 11-12-2021deposit of Omicron variant and compared with Delta and Alpha variants. Parts of the major differences were shown here to demonstrate major feature of Omicron having deletions at H69, V70, V143, Y144, Y145 amino acids but not in F167, R168 amino acid positions as found in Delta variants. L212 deletion was not aligned here and RE insertion will be EPE based on corona virus genome alignment (see, Figure) due to similarity of VR sequences (green circles) between omicron and Wuhan and Delta viruses. Corona virus patient samples were Nasal swab, Oral swab, Saliva and Mucus origin between 24th November 2021 to 5th December 2021 from United States (CA, MI, MA, GA, NY, AZ, AL, KS, CO, TX).

Figure 5: Multi-alignment of omicron corona virus genome (30kb) which confirmed a deletion of one amino acid (L212) following three amino acid (EPE) insertion at 215 position of spike protein as well as H69, V70, G143, Y144, Y145 deletions. Part of the genome was shown here.
Data analysis of dated 13-12-2021 was resulted three omicron isolates deposited by Lemieux JE, et al., but appeared all incomplete sequences (accession nos. OL823147 and OL823148 (USA, MA) and OL822906 (USA, NY). However, the same day deposited many sequences by Howard, et al., and Blankenship HM, et al., had no omicron virus but mostly AY.X and few B.1.1.617.2. Similarly data deposited on 16-12-2021 had no omicron variant (Howard D, et al., and Diagnostics G, et al.,) and for dated 17-12-2021 only two omicron selected (accession nos. OL890283 (Lemieux J, et al.) and OL901854 (Linares-Perdo DJ, et al.). Interestingly, twenty four omicron viruses were deposited on 18-12-2021 by Lemieux JE, et al., USA. Accession nos. were OL903123 (NH, 3-12-2021), OL880661(MA, 3-12-2021), OL902594 (CT, 4-12-2021), OL903509(MA, 5-12-2021), OL913145(RI, 6-12-2021), OL904534(MA, 6-12- 2021), (OL904790(MA, 6-12-2021) OL904791(MA, 6-12- 2021), OL903931(VT, 8-12-2021), OL904803(NH, 8-12-2021), OL904422(MA, 8-12-2021), OL904499(MA, 9-12-2021) and seven omicron data from Massachusetts with collection date 8-12-2021 and accession nos. OL903558, OL903553, OL903821, OL903690, OL903698, Ol903660, OL903604 and three from New York (accession nos. OL903848, OL903853, OL903850) and two from Rhodes Island (accession nos. OL903865, Ol903866). On the same day (18-12-2021) Nickerson DA, et al., (USA) was deposited four omicron variants with accession no. OL903977 (collection date 9-12-2021) and accession nos. OL903978/79/80 with collection date 12-12-2021. Most incomplete sequences were found at the AAs 303-393, 313-393, and 425-439 indicating primers would not worked during initial sequencing due to strong variations in the genome of omicron virus. Such incomplete S proteins were analyzed by Multalin software and the resulted with unique deletions and insertions for omicron (data not shown).
Data analyzed for day 20-12-2021 deposit by Howard D, et al., Linares-Perdo J, et al., and Grimaldo V, et al., appeared mostly AY.X and few B.1.617.2 . The data analysis for day 21-12-2021 deposit by Parrott T, et al., gave four omicron complete sequences with accession no OL960535/36/37/38 with collection date 8-12-2021. Same day, Graffin J, et al., deposited 28 omicron sequences from Minnosota, USA with accession nos. OL964103, OL964105/06, OL964108/09, OL964111/12/13/14/15, OL964117/18, OL964120/21/22/23, OL964125, OL964127/28/29/30/31/32, and OL964134/35/36/37/38. S proteins were partial and few only analyzed to confirm omicron signatures (data not shown). However, sequences deposited by Howard D, et al., Blankenship HM, et al., Pokhard A, et al., and Ritter J, et al., produced mostly AY.x delta corona virus (data not shown).
Data analyzed on 22-12-2021 deposit by Gohl DM, et al., Banu LA, et al., Irfan M, et al., Beukelman R, et al., Bankers L, et al., were mostly incomplete and multi-alignment produced some false image with no omicron sequence was interpreted. But Lemieux JE, et al., deposited six omicron sequences with accession nos. OL976589 (USA, MA), OL976472 (USA, MA), OL976989 (USA, ME), OL976899 (USA, MA), OL977025 (USA, VT), OL977069 (USA, VT)] in the same day with collection dates 7-12-2021 to 11-12-2021. Also Gener A, et al., deposited few complete omicron sequences from California with accession nos. OL977473 (4-12-2021), OL977502 (13-12-2021) as well as some incomplete omicron sequences with accession nos. OL977503 (13-12-2021), OL977504 (13-12-2021) and OL977661 (14-12-2021) (data not shown).
The omicron sequences were deposited by Lemieux JE, et al., from USA with accession nos. OL976589, OL976472, OL976899 (USA, MA) and OL977025, Ol977069 (USA, VT) and OL976989 (USA, ME) on dated 23-12-2021. Kandal S, et al., deposited one omicron sequence on 23-12-2021 with accession no. OL988626 (USA, AR; 10-12-2021). Howard D, et al., deposited on the same day many omicron sequences with accession numbers OL991113 (USA, OH), OL991168 (USA, TX), OL991171 (USA, WV) with collection date 1-12-2021 and OL991968 (USA, MD) with collection date 7-12-2021 and a partial omicron sequence with accession no. OL991968 (CA, 4-12-2021) (data not shown).
The last day of our analysis was dated 24-12-2021 deposited sequences. We found by multi-alignment that Nickerson, et al., deposited many omicron sequences originated in Washington with accession numbers OM003743 (19-12-2021), OM003730 (15-12- 20210, OMOM003721 (17-12-2021), OM003729/27/28/32 (15-12- 2021), OM003707/14/26/34 (13-12-2021), OM003719/35/37 (17- 12-2021), OM003711/17/23 (13-12-2021), OM003716 (15-12-2021) and OM003744/41/42 (19-12-2021). We also found Ryan KA of United Kingdom deposited one omicron sequence with accession number OM003685 (27-11-2021). On 24-12-2021, Howard D, et al., also deposited many omicron sequences originated in the New York with accession numbers OM005692, OM005638, OM005669, OM007728, OM007698, OM007702, OM007701 and OM007731 (collection dates 7-8th December, 2021). He also deposited many omicron sequences originated in the different US States with accession numbers OM007718, OM007685 (New Jersey, 8-12-2021), OM007637 (Maryland, 7-12-2021), OM007625 (Pennsylvania, 7-12- 2021), OM007696 (District of Columbia, 8-12-2021) and OM007970 (Hawaii, 13-12-2021) (data not shown).
We got all the 141 suspected 1270 aa length complete + incomplete omicron sequences and found 21 complete sequences. Multialignment produced three mutations (D212Y in UHO53537, R343K in UHO53468/91 and UHO53648 and A698V in UHO53131, UGO96815 and UGO96803) in the omicron sequences. So, up to 24th December 2021, the Database (December 6 to December 24, 2021 total sequences deposited were 3,71,307) penetration of complete + incomplete omicron sequences were 0.0379% and for complete sequences it further very reduced to 0.0056%. This indicated it was very hard to get omicron virus sequences which was complete and authentic (Figure 6).

Figure 6: Complete omicron corona virus spike protein sequences up to 24th December, 2021. Protein IDs and collection dates in 2021 were given and compared with Wuhan, Alpha and Delta corona virus spike proteins. Part of the alignment was given showing V143, Y144 and Y145 three amino acids deletions.
Interestingly, on the Christmas day (25-12-2021) Howard D, et al., deposited 216 omicron virus sequences to the NCBI Virus database indicating huge omicron virus transmission that likely competed the huge transmission of the delta corona virus. To confirm the omicron virus, we had selected few sequences from the different part of the NCBI multi-alignment graph and analyzed (Figure 7).

Figure 7: Data analysis of omicron sequences deposited on 25-12-2021 by Howard D, et al., USA. Only three sequences (OM01136, OM010507, OM011026) appeared complete and rest had ambiguity about three amino acids (EPE) insertions in the spike protein. Thus, although ~216 omicron virus sequences were deposited by Howard D, et al., on dated 25th December 2021, authentic omicron corona virus spike protein sequence was still hard to detect.
We want to learn more signatures for omicron corona virus. We found no changes in the furine cleavage site of S protein (Figure 8) (Hoffmann, et al., 2020). When we analyzed the N-protein sequences from omicron viruses, we detected unique three amino acid deletion (ERS) at position 30 and multi-alignment data presented in figure 9. Interestingly, N-protein had point mutations like P13L, R203K, G204R and in some D343G. Similarly, two extra new mutation in the M-protein were detected (D3G, A63T) (Figure 10) but no mutation in the ORF3a protein (data not shown). Small structural E protein (75 aa) in omicron virus has one mutation (T9I; data not shown). Never the less omicron virus transmission is rapidly increasing in 90 countries and in some US states it is about 10% where as in South Africa was 90% now. Thus, the rate of omicron transmission has increased in UK and Germany where as about 400 patients were detected in India. Death already was reported in England and USA although the omicron disease appeared to be less virulent where oxygen support and hospitalization were unnecessary.

Figure 8: Multi-alignment showing no mutation in the spike protein furine cleavage site of Wuhan, Alpha, Delta and Omicron variants of COVID-19.
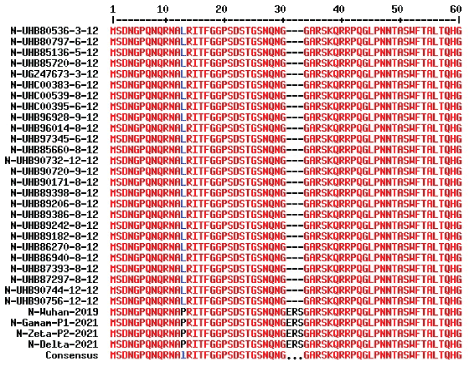
Figure 9: Three amino acids deletion of N-protein (416 aa) is an indicator of omicron variant corona virus. Data dated 18-12-2021 was analyzed and compared with other old variants. P13L, R203K, G204R point mutations were also detected.

Figure 10: Multi-alignment of M-protein of different variant of corona virus. D3G and A63T mutations in Omicron variant were detected.
Surprisingly, on 25-12-2021 huge omicron data deposited in the NCBI SARS-CoV-2 Database as we reported higher than 216 omicron sequences. But dated 27-12-2021 analysis we did not find the pattern of multi-alignment for omicron virus (Figure 1C). Ultimately we discovered the heavy read lines were found in Delta variants (reversed) due to huge deposition of omicron sequences from 25-12-2021 onwards. We then BLAST searched the 60 nt hyper-variable region of spike protein (22894 5’- AAC TGA AAT CTA TCA GGC CGG TAA CAA ACC TTG TAA TGG TGT TGC AGG TTT TAA TTG TTA3’ 22953) and we found 3815 possible omicron sequences instead of only four (4) found on dated 08-12-2021 BLAST search. Such data was astonishing and a huge spread of corona virus was evident in the USA from the second week of December, 2021. Dated 29-12-2021 and 31- 12-2021 analysis, we discovered a mixed trend of alignment suggesting our method still work well. Likely omicron penetration increased to 1.2% during end of December, 2021.
Next we analyzed the important of S gene mutations on corona virus diagnostics. Many RT-PCR kits utilized the S gene primers where some kits appeared unable to give RT-PCR data from the S gene region. We determined the sequence variation in omicron virus as compared to Wuhan 2019 strain. Data presented in figure 11 where two or more regions in the genome were presented. We made 10 primers set using NCBI Primer Design software and one was located in the S gene. Analysis found by BLAST that forward primer (F3= 5’-23518GAC TAA GTC TCA TCG GCG GG23537-3’ would not hybridize to omicron genome but reverse primer 5’-24130CCC ACA TGA GGG ACA AGG AC24111-3’ did well. Old primers designed for Wuhan strain were hardly identify S gene of omicron variants pinpointing new primers design were necessary to track omicron transmission using S gene primers. This is an example to be aware for RT-PCR using old primers for the detection of omicron virus spread. However, the primer pairs F8-5’- GGC AAA CCA CGC GAA CAA AT-3’ and R8 5’- GAG GGT CAA GTG CAC AGT CT-3’ (1145 bp; Tm=60oC) worked well for all Wuhan (accession no. NC_045512.2), alpha B.1.1.7 (accession no. OD984292), delta B.1.617.2 (accession no. OV104747) and omicron B.1.1.529 (accession no. OL672836) corona viruses. Similarly to sequence the hyper-variable point mutated region (300-550 AA) we devised a forward primer from F8 reverse primer region F9-5’-CTG TGC ACT TGA CCC TCT CTC-3’ and a reverse primer downstream (R9-5’-CAC GGA CAG CAT CAG TAG TGT-3’) giving a 863 bp DNA product (Tm=60oC) for all Wuhan, alpha, delta and omicron corona viruses.

Figure 11: Seq-2 BLAST similarity analysis between December 2019 Wuhan corona virus (accession no. NC_045512.2) and November 2021 omicron corona virus variant (accession no. OL721912). Only multiple nt. difference positions of spike gene due to mutation and deletion were presented here.
Interestingly, BLAST search of another part of hyper-variable region (22954 5’- CTT TCC TTT ACG ATC ATA TAG TTT CCG ACC CAC TTA TGG TGT TGG TCA CCA ACC ATA CAG-3’ 23013) resulted same three omicron sequences with 100% similarity but also we identified a 20nt exact match with chromosome-2 (nt. 13807869 to 13807888) of Seladonia tumulonum (bee) and 19 nt exact match with chromosome 16 (nt. 1796578 to 1796596) of Stemomphata cineraria (sea snail). It codes amino acids 488-493 (NH2-PLRSYS-CO2H) region of spike protein of omicron corona virus (Figure 12). We do not know why such similarity of viral sequence to lower eukaryotes genome like fly or mollusca! However, such information could have some interest to some evolutionary biologist.
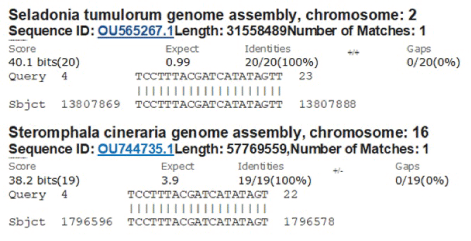
Figure 12: Detection of first perfect sequence homology of S gene with bee ch-2 and snail ch-16.
Then, we wanted know how six amino acids deletions and three amino acids insertions with many mutations could protect the omicron virus S protein functional and stable giving the capacity for higher transmission than alpha and delta variants? When we analyzed the amino acid composition of spike proteins, we found no gross changes in omicron as compared to Wuhan and Alpha strains. Data presented in figure 13 (www.expasy.org/cgi-bin /portparam). Very minor changes were noticed and boxed for Arginine, Aspergine, Glutamine, Phenyl Alanine and Serine (Figure 11). Acidic amino acids (Asp + Glu) and basic amino acids (Arg + Lys ) were found 110 and 103 in Wuhan virus, 109 and 103 respectively for B.1.1.7 alpha virus where as such amino acids in Omicron corona virus were found 111 for both acidic and basic amino acids. Alipathic index for Wuhan virus was found 84.67 and for omicron virus 84.95 and for alpha virus 84.65. Instability index for Wuhan, alpha and omicron viruses were found 33.01, 32.82 and 34.65 respectively explaining omicron corona virus spike protein was very stable. That may be the one of the cause of higher transmission of omicron corona virus due to stable interaction with ACE-2 receptor and S protein. However, the interaction of other >15 mutations in the RBD domain of omicron corona virus were unknown. Hydrophobic plot of Wuhan and omicron corona viruses were presented in figure 14. There are some differences as shown by green box.
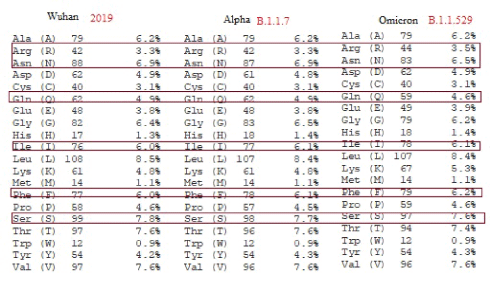
Figure 13: Difference in amino acids composition among Wuhan, Alpha and Omicron variants of corona virus (COVID-19).

Figure 14: Hydrophobicity plot showing minor differences (Boxed) between S protein of Wuhan and Omicron corona viruses.
Omicron variant of corona viruses are rapidly spreading in the world including USA, UK, Australia, and India. We analyzed here the NCBI database to conclude that the omicron virus was rapidly spreading in the United States of America and spike protein of omicron corona virus was very stable although it had more deletions and mutations than alpha and delta variants. Recently, a paper was published on African omicron corona virus phylogenetics using GISAIP database and the accession no of the first omicron virus genome (29684 bp) was EPI_ISL_6640916 with collection date 11-11-2021 and submission date 23-11-2021 [25]. However, I have no access to such database. The molecular mechanism of the new 26 unknown spike protein mutations were yet to know but such changes occurred with higher transmission and combined 6 deletions of amino acid surely increased immune escape [26]. We analyzed enormous data from NCBI SARS-CoV-2 database from November-December, 2021 and detected about ~141 omicron sequences (upto 24-12-2021 deposit). But such sequences were mostly incomplete and we finally got about 21 authentic omicron spike protein sequences (Figure 6). On Christmas day Howard D, et al., deposited about 216 omicron sequences but mostly incomplete due to ambiguity of insertion sequences but there were enough complete sequences for spike protein to analyze. I faced tremendous problem to get an authentic omicron spike protein sequence during last week of November, 2021 when omicron reports were mounting in the media. In January, 2022, the number of Omicron sequences were more than Delta variants!
Question arises such hyper-variable difference of omicron spike protein could be involved in the vaccine failure for the corona vaccine made from S gene! Analysis suggested that some hydrophobic regions had similarity and surely partial protection possible! Study indicated that only 20% and 24% of BNT162b2 vaccine recipients had detectable neutralizing antibody against the omicron variant HKU691 and HKU344-R346K, respectively, while none of the Coronavac recipients had detectable neutralizing antibody titre against either omicron isolate. Omicron variant escapes neutralizing antibodies elicited by BNT162b2 or Coronavac [27]. Using animal model Starr TN, et al., [28] showed that the antibodies, S2H97 and S2E12 bound with high affinity across all sarbecovirus clades to a cryptic epitope and prophylactically protects hamsters from viral challenge [28]. Further study suggested that E484K mutation evaded antibody neutralization elicited by infection or vaccination and further enhanced by K417N and N501Y mutations [29]. A very similar conclusion was confirmed by antibodies raised against deletion mutants of RBD domain of spike protein [30]. Wang R, et al., [20,21] and others (2021) showed that the South African variant B.1.351 was the most resistant to current monoclonal antibodies and convalescent plasma from Wuhan virus infected individuals, followed by United Kingdom alpha variant (B.1.1.7) and the Brazilian gamma variant (P.1) with Y144del and 242-244del mutations and important D614G and N501Y mutations as well as K417N/T, E484K/Q mutations in the RBD domain of the spike protein of SARS-CoV-2 [27,31,32] (Wand, et al., 2021). Another study indicated that neutralizing antibodies elicited by inactivated corona virus vaccine and RBD-subunit spike protein vaccine against B.1.617 and B.1.1.7 variants enhanced viral entry and membrane fusion, as well as more resistant to antibody neutralization [33-35]. Thus, all natural mutations have allosteric effects that drive either interspecies transmission or escape from antibody neutralization [36]. Omicron virus was already transmitted in 90 countries and likely will be threat to humanity [37]. Omicron may be ten times more contagious than the original virus and twice more infectious than delta variant. We have shown that omicron viruses are greatly affected many US States including CA, NY, CO, MN and NJ. Further, omicron may be twice more likely to escape current vaccines than the delta variant [38]. Wang R, et al., identified fast-growing RBD mutations like N439K, S477N, S477R, and N501T that enhanced the RBD and ACE2 binding. L452R mutation in the spike reduces its interaction with Wuhan corona virus antibodies. Similarly, mutations E484K and K417N found in South Africa and L452R and E484Q found in India variants could be responsible for such reduced antibody interaction [21]. Miller NL, et al., preprint disclosed that the omicron variant increased antibody escape due to mutations in class 3 and 4 antibody epitopes in the spike protein as well as enhanced transmissibility via disruption of ligand-receptor interface [26]. Finally, molecular biology of omicron virus has just started to define its functions of genetic changes [25]. Although mild symptoms of fever, cold and pneumonia reported, delmicron (Delta + Omicron) has created a havoc calamity in the world [39]. Remdesivir drug however has some benefit to control corona virus spread targeting RNA dependent RNA polymerase as well as some immune drug were discovered [40]. We have pinpointed the differences in the spike protein of omicron but omicron spike protein appeared very stable to interact with ACE-2 receptor. But genomic mutations may affect RT-PCR (Figure 11) and thus new RTPCR primers were presented from conserved regions. Interestingly, now more sequences for omicron virus will be available in the NCBI SARS-CoV-2 Database.
The work was not funded by any agency. The author thanks CDC and NCBI for the data provided. AKC is a retired professor.
Download Provisional PDF Here
Aritcle Type: RESEARCH ARTICLE
Citation: Chakraborty AK (2022) Hyper-Variable Spike Protein of Omicron Corona Virus and Its Differences with Alpha and Delta Variants: Prospects of RT-PCR and New Vaccine. J Emerg Dis Virol 7(1): dx.doi.org/10.16966/2473-1846.166
Copyright: © 2022 Chakraborty AK. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access