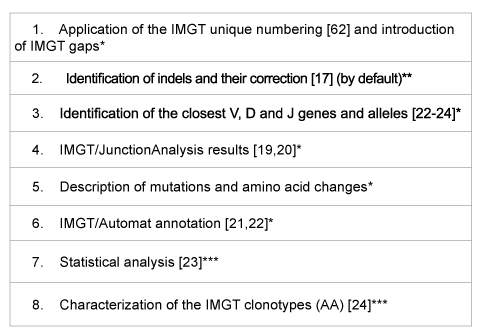
Table 1: IMGT/HighV-QUEST functionalities and results
*common to IMGT/HighV-QUEST and IMGT/V-QUEST (same algorithm
and same IMGT reference directories).
**option in IMGT/V-QUEST.
***specific to IMGT/HighV-QUEST.
Véronique Giudicelli Patrice Duroux Arthur Lavoie Safa Aouinti Marie-Paule Lefranc* Sofia Kossida*
IMGT®, the international ImMunoGenetics information system®, Laboratoire d’ImmunoGénétique Moléculaire LIGM, Institut de Génétique Humaine IGH, UPR CNRS 1142, Montpellier University, Montpellier, France*Corresponding author: Marie-Paule Lefranc and Sofia Kossida, IMGT®, the international ImMunoGenetics information system®, Laboratoire d’ImmunoGénétique Moléculaire LIGM, Institut de Génétique Humaine IGH, UPR CNRS 1142, Montpellier University, 141 rue de la Cardonille, 34396 Montpellier cedex 5; Tel.: 33-434-359965; Fax: 33-434-359901; E-mail: Marie-Paule.Lefranc@igh.cnrs.fr, Sofia.Kossida@igh.cnrs.fr
The analysis of adaptive immune repertoires in autoimmune and infectious diseases represents a fascinating challenge because of the natural huge diversity of the antigen receptors that are the immunoglobulins or antibodies and T cell receptors, expressed by B cells and T cells, respectively. The high throughput analysis resulting from the next generation sequencing technologies offers an invaluable large scale characterization of the expressed repertoires and, consequently, a better understanding of the protective and pathogenic immune responses. IMGT®, the international ImMunoGeneTics information system® (Centre National de la Recherche Scientifique and Montpellier University) is the global reference in immunogenetics and immunoinformatics. By its creation in 1989, IMGT® marked the advent of immunoinformatics, which emerged at the interface between immunogenetics and bioinformatics. IMGT® is specialized in the immunoglobulins or antibodies, T cell receptors, major histocompatibility proteins of humans and other vertebrate species, and in the proteins of the immunoglobulin superfamily and major histocompatibility superfamily of any species. IMGT® has been built on the IMGT-ONTOLOGY axioms and concepts, which bridged the gap between genes, sequences and three-dimensional structures. The concepts include the IMGT® standardized keywords (identification), IMGT® standardized labels (description), IMGT® standardized nomenclature (classification), IMGT unique numbering and IMGT Colliers de Perles (numerotation). IMGT® comprises seven databases, 15,000 pages of web resources and 17 tools, and includes data from fishes to humans, for basic to veterinary, medical and translational research. IMGT/HighV-QUEST, the high throughput version of IMGT/V-QUEST and IMGT/Junction Analysis, created in 2010, is the first and, so far, the only online portal for analysis of the immunoglobulin and T cell receptor repertoires obtained from next generation sequencing. High-quality results contribute to the large scale vision of antigen receptor repertoires and understanding of the adaptive immune responses (protective in vaccination, cancer and infections and pathogenic in autoimmunity) for diagnosis, prognosis and therapy.
IMGT; Immunogenetics; Immunoinformatics; IMGT-ONTOLOGY; Immunoglobulin (IG); T cell receptor (TR); Immune repertoire; IMGT unique numbering; Next generation sequencing
IMGT®, the international ImMunoGeneTics information system® [1,2], was created in 1989 by Marie-Paule Lefranc at Montpellier, France (CNRS and Montpellier University). The founding of IMGT® marked the advent of immunoinformatics, a new science, which emerged at the interface between immunogenetics and bioinformatics. For the first time, immunoglobulin (IG) or antibody and T cell receptor (TR) variable (V), diversity (D), joining (J) and constant (C) genes were officially recognized as “genes” as well as the conventional genes [3-6]. This major breakthrough allowed genes and data of the complex and highly diversified adaptive immune responses to be managed in genomic databases and tools.
The adaptive immune response was acquired by jawed vertebrates (or gnathostomata) more than 450 million years ago and is found in all extant jawed vertebrate species from fishes to humans. It is characterized by a remarkable immune specificity and memory, which are properties of the B and T cells owing to an extreme diversity of their antigen receptors. The specific antigen receptors comprise the IG or antibodies of the B cells and plasmacytes [3], and the TR [4]. The IG recognizes antigens in their native (unprocessed) form, whereas the TR recognizes processed protein antigens, which are presented as peptides by the highly polymorphic major histocompatibility (MH, in humans HLA for human leucocyte antigens) proteins.
The potential antigen receptor repertoire of each individual is estimated to comprise about 2 × 1012 different IG and TR, and the limiting factor is only the number of B and T cells that an organism is genetically programmed to produce [3,4]. This huge diversity results from the complex molecular synthesis of the IG and TR chains and, more particularly, of their variable domains (V-DOMAIN) which, at their N-terminal end, recognize and bind the antigens [3,4]. The IG and TR synthesis includes several unique mechanisms that occur at the DNA level: combinatorial rearrangements of the V, D and J genes that code the V-DOMAIN (the V-(D)-J being spliced to the C gene that encodes the C-REGION in the transcript); exonuclease trimming at the ends of the V, D and J genes and random addition of nucleotides by the DNA nucleotidylexotransferase (DNTT, terminal deoxynucleotidyl transferase, TdT) that creates the junctional N-diversity regions, and later during B cell differentiation, for the IG, somatic hypermutations, gene conversion (e.g., in birds), and class or subclass switch in higher vertebrates [3,4].
IMGT® manages the diversity and complexity of the IG and TR and the polymorphism of the MH of humans and other vertebrates. IMGT® is also specialized in the other proteins of the immunoglobulin superfamily (IgSF) and MH superfamily (MhSF) and related proteins of the immune system (RPI) of vertebrates and invertebrates [2]. IMGT® provides a common access to standardized data from genome, proteome, genetics, and two-dimensional (2D) and three-dimensional (3D) structures. IMGT® is the acknowledged high-quality integrated knowledge resource in immunogenetics for exploring immune functional genomics. IMGT® comprises seven databases (for sequences, genes, and 3D structures) [7-12] and 17 online tools [13-28], as well as more than 15,000 pages of web resources (e.g., IMGT Scientific chart, IMGT Repertoire, IMGT Education > Aide-mémoire [29], the IMGT Medical page, the IMGT Veterinary page, the IMGT Biotechnology page, the IMGT Immunoinformatics page) [2]. IMGT® is the global reference in immunogenetics and immunoinformatics [30-45]. Its standards have been endorsed by the World Health Organization-International Union of Immunological Societies (WHOIUIS) Nomenclature Committee since 1995 (first IMGT® online access at the 9th International Congress of Immunology, San Francisco, CA, USA) [46,47] and the WHO International Nonproprietary Names (INN) Programme [48,49].
The accuracy and the consistency of the IMGT® data are based on IMGT-ONTOLOGY [50-52], the first, and so far unique, ontology for immunogenetics and immunoinformatics [50-69]. IMGT-ONTOLOGY manages the immunogenetics knowledge through diverse facets that rely on seven axioms: IDENTIFICATION, DESCRIPTION, CLASSIFICATION, NUMEROTATION, LOCALIZATION, ORIENTATION, and OBTENTION [51,52,56]. The concepts generated from these axioms led to the elaboration of the IMGT® standards that constitute the IMGT Scientific chart: e.g., IMGT® standardized keywords (IDENTIFICATION) [57], IMGT® standardized labels (DESCRIPTION) [58], IMGT® standardized gene and allele nomenclature (CLASSIFICATION) [59], IMGT unique numbering [60-65] and its standardized graphical 2D representation or IMGT Colliers de Perles [66-69] (NUMEROTATION).
With a focus on IG and TR immune repertoires, we first review the fundamental information generated from these IMGT-ONTOLOGY concepts which led to the IMGT Scientific chart rules, and then IMGT/ HighV-QUEST [18,23,24], the online web portal for the analysis of rearranged IG and TR repertoire nucleotide sequences from next generation sequencing (NGS). IMGT/HighV-QUEST is the high throughput version of IMGT/V-QUEST [13-18] with the results of the integrated IMGT/ JunctionAnalysis [19,20] and of IMGT/Automat [21,22]. IMGT/HighVQUEST runs against IMGT reference directories built from sequences annotated in IMGT/LIGM-DB [7], the IMGT® nucleotide database (176,948 sequences from 347 species in July 2015), and from IMGT/GENE-DB [8], the IMGT® gene database (3567 genes and 5258 alleles from 22 species, of which there were 715 genes and 1469 alleles for Homo sapiens and 869 genes and 1319 alleles for Mus musculus in July 2015).
The unified IMGT® approach is of major interest for bridging knowledge from IG and TR repertoires in physiological and pathological situations [70-75], for guiding antibody discovery, humanization and engineering [76-86], and providing the same standardized results for NGS repertoires in autoimmune and infectious diseases.
More than 325 IMGT® standardized keywords (189 for sequences and 137 for 3D structures) were precisely defined [57]. They represent the controlled vocabulary assigned during the annotation process and allow standardized search criteria for querying the IMGT® databases and for the extraction of sequences and 3D structures. They have been entered in BioPortal [87] at the National Center for Biomedical Ontology (NCBO) in 2010.
Standardized keywords are assigned at each step of the molecular synthesis of an IG. Those assigned to a nucleotide sequence are found in the “DE” (definition) and “KW” (keyword) lines of the IMGT/ LIGM-DB files [7]. They characterize, for instance, the gene type, the configuration type and the functionality type [57]. There are six gene types: variable (V), diversity (D), joining (J), constant (C), conventional-withleader, and conventional-without-leader. Four of them (V, D, J, and C) identify the IG and TR genes and are specific to immunogenetics. There are four configuration types: germline (for the V, D, and J genes before DNA rearrangement), rearranged (for the V, D, and J genes after DNA rearrangement), partially-rearranged (for D gene after only one DNA rearrangement) and undefined (for the C gene and for the conventional genes, which do not rearrange). The functionality type depends on the gene configuration. The functionality type of genes in germline or undefined configuration is functional (F), open reading frame (ORF) or pseudogene (P). The functionality type of genes in rearranged or partially-rearranged configuration is either productive (no stop codon in the V-(D)-J region and in-frame junction) or unproductive (stop codon (s) in the V-(D)-J region, and/or out-of-frame junction).
The 20 usual amino acids (AA) have been classified in 11 IMGT physicochemical classes (IMGT®[1], IMGT Education > Aide-mémoire > Amino acids). The amino acid changes are described according to the hydropathy (three classes), volume (five classes) and IMGT physicochemical classes (11 classes) [29]. For example Q1 > E (+ + −) means that in the amino acid change (Q > E), the two amino acids at codon 1 belong to the same hydropathy (+) and volume (+) classes but to different IMGT physicochemical properties (−) classes [29]. Four types of AA changes are identified in IMGT®: very similar (+ + +), similar (+ + −, + − +), dissimilar (− − +, − + −, + − −), and very dissimilar (− − −).
More than 560 IMGT® standardized labels (277 for sequences and 285 for 3D structures) were precisely defined [58]. They are written in capital letters (no plural) to be recognizable without creating new terms. Standardized labels assigned to the description of sequences are found in the “FT” (feature) lines of the IMGT/LIGM-DB files [7]. Querying these labels represent a big plus compared to the generalist databases (GenBank/European Nucleotide Archive (ENA)/DNA Data Bank of Japan (DDBJ)). Thus it is possible to query for the “CDR3-IMGT” of the human rearranged productive sequences of IG-Heavy-Gamma (e.g., 1828 CDR3- IMGT obtained, with their sequences at the nucleotide or amino acid level). The core labels include V-REGION, D-REGION, J-REGION, and C-REGION which correspond to the coding region of the V, D, J and C genes. IMGT structure labels for IG chains and domains allow describing the chain and domains of 3D structures (see for example http://www.imgt. org/3Dstructure-DB/cgi/details.cgi?pdbcode=1HZH). Correspondence between human IG structure labels and sequence labels, links and correspondence between standardized keywords for IDENTIFICATION and standardized labels for DESCRIPTION in IMGT/PROTEIN-DB and IMGT/3Dstructure-DB are detailed in IMGT Scientific chart (see http://www.imgt.org/IMGTScientificChart/SequenceDescription/ IMGT3Dkeywords.html). These labels are necessary for a standardized description of the IG sequences and structures in databases and tools [58].
Highly conserved amino acids at a given position in a domain have IMGT labels [58]. Thus three amino acid labels are common to the V and C domains: 1st-CYS (cysteine C at position 23), CONSERVEDTRP (tryptophan W at position 41), and 2nd-CYS (C at position 104) [60-63,65]. Two other labels are characteristic of the V-DOMAIN and correspond to the first amino acid of the canonical F/W-G-X-G motif (where F is phenylalanine, W tryptophan, G glycine, and X any amino acid) encoded by the J-REGION: J-PHE or J-TRP (F or W at position 118) [60- 62,65].
The IMGT-ONTOLOGY CLASSIFICATION axiom was the trigger of immunoinformatics birth [45]. Indeed the IMGT® concepts of classification allowed us, for the first time, to classify the antigen receptor genes (IG and TR) for any locus (e.g., immunoglobulin heavy (IGH), T cell receptor alpha (TRA)), for any gene configuration (germline, undefined, or rearranged) and for any species (from fishes to humans). In higher vertebrates, there are three IG major loci (other loci correspond to chromosomal orphon sets, genes of which are orphons, not used in the IG chain synthesis). The IG major loci include the immunoglobulin heavy (IGH), and for the light chains, the immunoglobulin kappa (IGK) and the immunoglobulin lambda (IGL) in higher vertebrates and the immunoglobulin iota (IGI) in fishes (IMGT®[1], IMGT Repertoire).
Since the creation of IMGT® in 1989, at New Haven during the 10th Human Genome Mapping Workshop (HGM10), the standardized classification and nomenclature of the IG and TR of humans and other vertebrate species have been under the responsibility of the IMGT Nomenclature Committee (IMGT-NC).
IMGT® gene and allele names are based on the concepts of classification of “Group”, “Subgroup”, “Gene” and “Allele” [59]. “Group” allows classification of a set of genes that belong to the same multigene family, within the same species or between different species. For example, there are 10 groups for the IG of higher vertebrates: IGHV, IGHD, IGHJ, IGHC, IGKV, IGKJ, IGKC, IGLV, IGLJ, and IGLC. “Subgroup” allows classification of a subset of genes that belong to the same group and that, in a given species, share at least 75% identity at the nucleotide level, e.g., Homo sapiens IGHV1 subgroup. Subgroups, genes, and alleles are always associated to a species name. An allele is a polymorphic variant of a gene, which is characterized by the mutations of its sequence at the nucleotide level, identified in its core sequence, and compared to the gene allele reference sequence, designated as allele *01. For example, Homo sapiens IGHV1- 2*01 is the allele *01 of the Homo sapiens IGHV1-2 gene that belongs to the Homo sapiens IGHV1 subgroup which itself belongs to the IGHV group. For the IGH locus, the constant genes are designated by the letter (and eventually number) corresponding to the encoded isotypes (IGHM, IGHD, IGHG3…), instead of using the letter C. IG and TR genes and alleles are in capital letters whatever the species and are not italicized in publications. IMGT-ONTOLOGY concepts of classification have been entered in the NCBO BioPortal [87].
The IMGT® IG and TR gene names [3-6] were approved by the Human Genome Organization (HUGO) Nomenclature Committee (HGNC) in 1999 [88,89] and were endorsed by the WHO-IUIS Nomenclature Subcommittee for IG and TR [46,47]. The IMGT® IG and TR gene names are the official international reference and, as such, have been entered in IMGT/GENE-DB [8], in the Genome Database (GDB) [90], in LocusLink at the National Center for Biotechnology Information (NCBI) USA [91], in Entrez Gene (NCBI) when this database (now designated as “Gene”) superseded LocusLink [92], in NCBI MapViewer, in Ensembl at the European Bioinformatics Institute (EBI) [93], and in the Vertebrate Genome Annotation (Vega) Browser [94] at the Wellcome Trust Sanger Institute (UK). HGNC, Gene NCBI, Ensembl, and Vega have direct links to IMGT/GENE-DB [8]. IMGT® human IG and TR genes were also integrated in IMGT-ONTOLOGY on the NCBO BioPortal and, on the same site, in the HUGO ontology and in the National Cancer Institute (NCI) Metathesaurus. The Mus musculus IG and TR genes were provided in 2005 to the Mouse Genome Database (MGD) [95], following the IMGT® presentation of the 7 mouse IG and TR loci at the 19th International Mouse Genome Conference (IMGC) in Strasbourg, France. Amino acid sequences of human IG and TR constant genes (e.g., Homo sapiens IGHM, IGHG1, IGHG2) were provided to UniProt [96] in 2008. Since 2007, IMGT® IG gene and allele names have been used for the description of the therapeutic monoclonal antibodies (-mab suffix) and fusion proteins for immune applications (FPIA) (-cept suffix) of the WHO-INN programme [48,49].
The IMGT-ONTOLOGY NUMEROTATION axiom is acknowledged as the “IMGT® Rosetta stone” that has bridged the biological and computational spheres in bioinformatics [38]. The IMGT® concepts of numerotation comprise the IMGT unique numbering [60-65] and its graphical 2D representation, the IMGT Collier de Perles [66-69]. Developed for and by the “domain”, these concepts integrate sequences, structures, and interactions into a standardized domain-centric knowledge for functional genomics. The IMGT unique numbering has been defined for the variable V domain (V-DOMAIN of the IG and TR, and V-LIKEDOMAIN of IgSF other than IG and TR) [60-62], the constant C domain (C-DOMAIN of the IG and TR, and C-LIKE-DOMAIN of IgSF other than IG and TR) [63] and the groove G domain (G-DOMAIN of the MH, and G-LIKE-DOMAIN of MhSF other than MH) [64]. Thus the IMGT unique numbering and IMGT Collier de Perles provide a definitive and universal system across species, including invertebrates, for the sequences and structures of the V, C and G domains of IG, TR, MH, IgSF and MhSF [65,69,84].
IMGT/HighV-QUEST [23], created in October 2010, is the high throughput version of IMGT/V-QUEST. It is so far the only online tool available on the Web for the direct analysis of complete IG and TR V-DOMAIN nucleotide sequences from NGS. IMGT/HighV-QUEST analyzes up to 500,000 sequences per run and performs statistical analysis on the results [23,24], with the same degree of resolution and high-quality results as IMGT/V-QUEST [13-18,70] and IMGT/JunctionAnalysis [19,20]. Indeed IMGT/HighV-QUEST uses the same algorithm and runs against the same IMGT reference directory. In July 2015, more than 6.62 billions of sequences were analyzed by IMGT/HighV-QUEST [18,23,24], by 1157 users from 41 countries (44% users from USA, 37% from EU, 19% from the remaining world).
IMGT/HighV-QUEST functionalities are basically the same as IMGT/ V-QUEST [13-18], the IMGT® online tool for the analysis of nucleotide sequences of the IG and TR V-DOMAIN (Table 1). IMGT/HighV-QUEST numbers the user sequences according to the IMGT unique numbering [62] and introduces gaps accordingly. It identifies the variable (V), diversity (D) and junction (J) genes in rearranged IG and TR sequences and, for the IG, characterizes the nucleotide (nt) mutations and amino acid (AA) changes resulting from somatic hypermutations [22,24] by comparison with the IMGT/V-QUEST reference directory. The tool integrates IMGT/ JunctionAnalysis [19,20] for the detailed characterization of the V-D-J or V-J junctions, IMGT/Automat [21,22] for a complete sequence annotation with the delimitation of the IMGT labels of description. By default IMGT/ HighV-QUEST identifies the insertions/deletions (indels) which are NGS errors resulting from homopolymer hybridization and corrects them [24]. Statistical analysis and characterization of the IMGT clonotypes (AA) [24] are functionalities specific to IMGT/HighV-QUEST (Table 1).
The IMGT/HighV-QUEST uses the same reference directory as IMGT/ V-QUEST [2]. It is organized per species and per locus and comprises the IMGT reference sequences. It includes the germline V, D and J nucleotide sequences of the core regions (V-REGION, D-REGION, and J-REGION) from all functional (F) genes and alleles, all open reading frame (ORF) and all in-frame pseudogenes (P) alleles from IMGT/GENE-DB [8]. It corresponds to the “F+ORF+ in-frame P” set, selected by default in “Advanced parameters”. The other options allow to exclude pseudogenes from the reference directory (“F+ORF”) or, for genomic analyses for example, to include orphon genes localized outside the main locus (“F+ORF including orphons” or “F+ORF+ in-frame P including orphons”).
By definition, the IMGT/V-QUEST reference directory contains one reference sequence for each allele. By default, the user sequences are compared with all alleles of the genes of the selected set. However, the “Advanced parameters” option “With allele *01 only” is useful if the user sequences need to be compared with different genes; and/or if the user sequences that use the same gene need to be aligned together (independently of the allelic polymorphism).
The IMGT/V-QUEST reference directories have been set up for species which are extensively studied, such as human and mouse for which a definitive gene nomenclature has been defined. This also holds true for the other species or taxons with incomplete IMGT reference directory sets with provisional nomenclature.
Each IMGT/V-QUEST programme version used by IMGT/HighVQUEST is identified by a number with a date (list of updates and modifications in the IMGT/V-QUEST programme versions page at http:// www.imgt.org/IMGT_vquest/share/textes/programversions.html). Each IMGT/V-QUEST reference directory used by IMGT/HighV-QUEST is similarly identified by a number with a date (list of changes in the IMGT/ V-QUEST reference directory releases page at http://www.imgt.org/ IMGT_vquest/share/textes/datareleases.html).
The IMGT/HighV-QUEST own version number reflects the updates of the functionalities that are specific to the system.
As for the other IMGT® databases and tools, IMGT/HighV-QUEST is freely available for academics. However, the IMGT/HighV-QUEST Welcome page requires user identification and provides, for new users, a link to register. User identification has been set to avoid non-relevant use and overload of the server, and to contact the user if needed. The user identification gives access to the IMGT/HighV-QUEST Search page (Figure 1).
The IMGT/HighV-QUEST Search page allows launching the analysis of sequences batches. The menu bar at the top of the page recalls the Login and provides a link to “Analysis history” to follow the process of the batches and to download the results (the other two menus, “Launch statistics” to perform statistics on sequences batch results, and “Statistics history” for the downloading of the corresponding statistics results, are not detailed here).
In the top part of the IMGT/HighV-QUEST Search page, the users have to provide a title for the analysis, to select the species (e.g., Homo sapiens) and the receptor type (IG or TR) or locus (e.g., IGL) from the drop down list. The file to be uploaded should contain IG or TR rearranged nucleotide sequences in FASTA and must be formatted in text. Users can choose to receive an e-mail notification when the analysis is queued in the local analysis queue and/or when the analysis is submitted for computer processing and/or when the analysis is completed and/or when the results can be downloaded and/or 5 days before the analysis is expired.
In Display results, the users can select the results files for the outputs: A. “Detailed View” (individual results file for each sequence) which is optional and only available for batches containing fewer than 150,000 sequences. B. “Files in CSV” which comprises up to eleven files if all are selected (the options are equivalent to those of the IMGT/V-QUEST “Excel files”).
In Advanced parameters, the users can choose parameters (identical to those of IMGT/V-QUEST). “Search for insertions and deletions” (an option in IMGT/V-QUEST) is selected by default in IMGT/HighVQUEST.
The results obtained from IMGT/HighV-QUEST are fully identical to those from IMGT/V-QUEST provided that the sequences have been processed with the same IMGT/V-QUEST programme version, the same IMGT/V-QUEST reference directory release, with an identical selection of species and receptor type or locus, and with the same options chosen in advanced parameters. The results are provided in a .txz archive containing a main folder with the eleven CSV files (if all were selected), and may include one subfolder with individual files, in text, for each submitted sequence (if the option “Include individual result files” of “Detailed View” was selected in the IMGT/HighV-QUEST Search Page). Text and CSV formats have been chosen in order to facilitate statistical studies for further interpretation and knowledge extraction. The format of the IMGT/HighV-QUEST results is the same as that of the option “Download in a zip archive” of the “Excel file” display of IMGT/V-QUEST.
The content of the eleven CSV files is summarized in Table 2. The CSV files contain one line per analysed sequence, and together may comprise up to 539 columns for a complete results report.
1. The “Summary” file #1 (29 columns with “Search for insertions and deletions” by default, or 25 without it) provides the synthesis of the analysis, i.e. the sequence names, the sequence functionality, the names of the closest V, D and J genes and alleles, the alignment scores and identity percentage for V and J, the D-REGION reading frame, the FR-IMGT and CDR-IMGT lengths, the amino acid (AA) JUNCTION (with frameshift(s) shown by # if any) provided by IMGT/JunctionAnalysis, the description of insertions and deletions if any, the warnings related to identification of the V and J gene and alleles and the sequence functionality, the orientation (“-” for opposite or “+” for direct) of the sequence at the submission, and the nucleotide sequence in the direct orientation with insertions in capital letters if any. The results of the IMGT/HighV-QUEST “Summary” file #1 are equivalent to those of the IMGT/V-QUEST “Results summary” (Figure 2).

Table 1: IMGT/HighV-QUEST functionalities and results
*common to IMGT/HighV-QUEST and IMGT/V-QUEST (same algorithm
and same IMGT reference directories).
**option in IMGT/V-QUEST.
***specific to IMGT/HighV-QUEST.
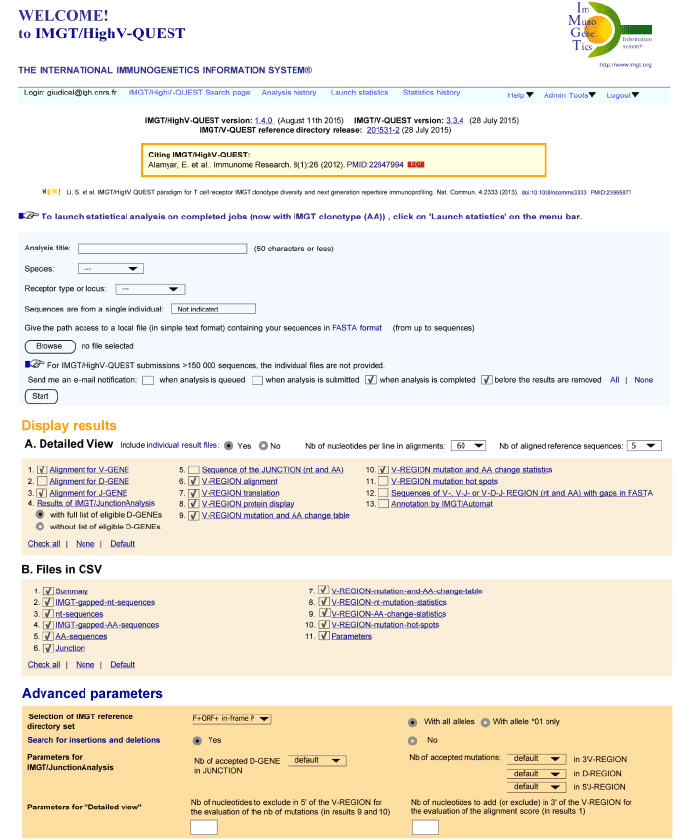
Figure 1. IMGT/HighV-QUEST Search page.
2. The “IMGT-gapped-nt-sequences” file #2 (18 columns) includes the nucleotide (nt) sequences gapped according to the IMGT unique numbering for the labels V-D-J-REGION, V-J-REGION, V-REGION, FR1-IMGT, CDR1-IMGT, FR2-IMGT, CDR2-IMGT, FR3-IMGT, and the nucleotide sequences of CDR3-IMGT, JUNCTION, J-REGION and FR4- IMGT. The “Nt-sequences” file #3 (66, 81, 102 or 114 columns, depending on the number of D (0, 1, 2 or 3) identified) includes all labels (ungapped nt sequences) delimited automatically by IMGT/Automat. The “IMGTgapped-AA-sequences” file #4 (18 columns) and the “AA-sequences” file #5 (18 columns) includes the AA sequences for the same labels as file #2, gapped (according to the IMGT unique numbering) and ungapped, respectively. The results of the files #2 to #5 are equivalent to those illustrated in IMGT/VQUEST “7. V-REGION translation” (Figure 3A) and “13. Annotation by IMGT/Automat” (Figure 3B).
3. The “Junction” file #6 includes all results of IMGT/JunctionAnalysis (40 columns for IGL, IGK, TRA and TRG sequences (no D), 53 (if one D), 73 (if two D) or 84 (if 3 D) columns for IGH, TRB and TRD sequences). All labels described in the junction are included with their nucleotide sequence and length (number of nt), as well as the number of trimmed nt for the 3’V-REGION, 5’ and 3’ D-REGION and 5’ J-REGION, the translation of the CDR3-IMGT and the translation of the JUNCTION (which includes 2nd-CYS (C104) and J-PHE (F118) or J-TRP (W118)), with frameshifts indicated by a # if any and without frameshift. The results of the “Junction” file #6 are equivalent to those of IMGT/V-QUEST “4. Results of IMGT/JunctionAnalysis” (Figure 4).
4. The “V-REGION-mutation-and-AA-change-table” file #7 (11 columns) includes the list of nt mutations and AA changes for the V-REGION, FR1- IMGT, CDR1-IMGT, FR2-IMGT, CDR2-IMGT, FR3-IMGT and CDR3- IMGT. The results of file #7 are equivalent to those of IMGT/V-QUEST “9. V-REGION mutation and AA change table” (Figure 5). The nt mutations and the corresponding AA changes for nonsilent mutations, are described for each FR-IMGT and CDR-IMGT, with their nt and codon position according to the IMGT unique numbering [62] and for the AA changes according to the IMGT AA classes [29] (IMGT Education > Aidemémoire).
5. The “V-REGION-nt-mutation-statistics” file #8 (130 columns) and the “V-REGION-AA-change-statistics” file #9 (109 columns) report the statistics of the nt mutations and AA changes, respectively. File #8 includes the total number of mutations, the number of silent and nonsilent mutations, the number of transitions and transversions. File #9 includes the number of AA changes according to AA class Change Type (+++, ++-, +-+, +--, -+-, --+, ---), and the number of AA class changes according to AA class Similarity Degree (Nb of Very similar, Nb of Similar, Nb of Dissimilar, Nb of Very dissimilar). In both files the results are given for V-REGION, FR1-IMGT, CDR1-IMGT, FR2-IMGT, CDR2-IMGT, FR3- IMGT and CDR3-IMGT. The results of files #8 and #9 are equivalent respectively to those of “Nucleotide (nt) mutations” and “Amino acids (AA) changes” of IMGT/V-QUEST “10. V-REGION mutation and AA change statistics” (Figure 6).
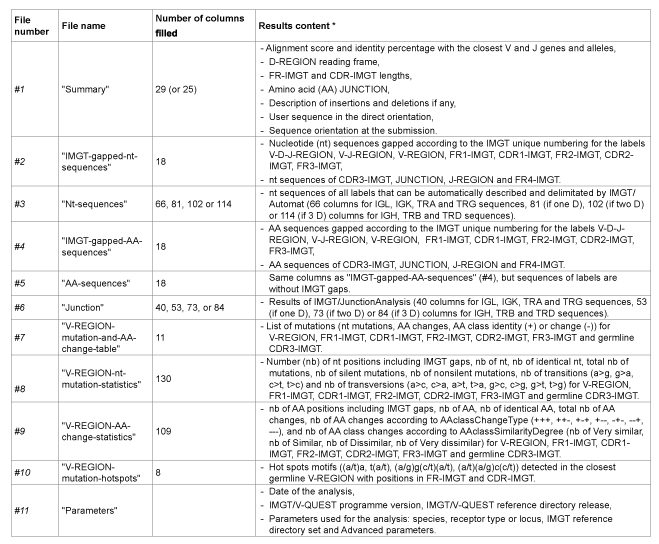
Table 2: Content of the eleven IMGT/HighV-QUEST results files in CSV format (results equivalent to those of the “Excel file” of IMGT/V-QUEST online). *: Files #1 to #10 comprise systematically sequence identification, i.e. the sequence name, the functionality, the names of the closest V gene and allele, and files #1 to #6 also include the D and J genes and alleles. The files #7 to #10 that report the analysis of mutations are used mostly for immunoglobulins (IG). Files #1 to #10 include one line per submitted sequence.
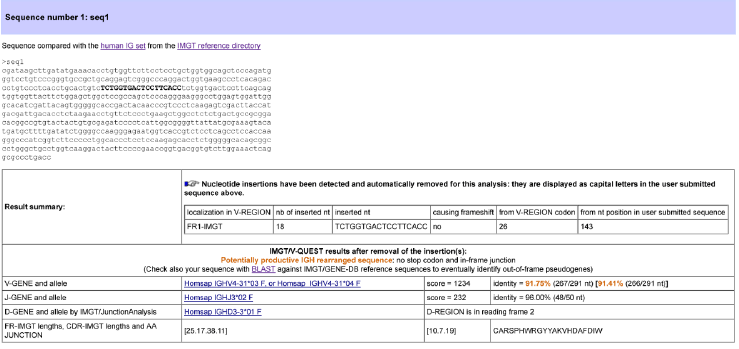
Figure 2. The results of the IMGT/HighV-QUEST “Summary” file #1 (29 columns) are equivalent to those of the IMGT/V-QUEST “Result summary table” with the option “Search for insertions and deletions” shown here (seq1: accession number AF013616 of IMGT/LIGM-DB).


Figure 3. The results of the IMGT/HighV-QUEST files #2, #4 and #5 (18 columns each) and file #3 (66, 81, 102 or 114 columns, depending on the number of D (0, 1, 2 or 3) identified) are equivalent to the results of IMGT/V-QUEST “7. V-REGION translation” (A) and “13. Annotation by IMGT/ Automat” (B) shown here (seq 2: accession number AB012909 of IMGT/LIGM-DB).

Figure 4. The results of the IMGT/HighV-QUEST “Junction” file #6 (40, 53, 73 or 84 columns, depending on the number of D (0, 1, 2 or 3) identified) are equivalent to those of IMGT/V-QUEST “4. Results of IMGT/JunctionAnalysis” shown here. Numbers of trimmed nt (dots in “Analysis of the JUNCTION”, here) are indicated in dedicated columns of the CSV file, for example in “3’V-REGION trimmed-nt nb”, “5’D-REGION trimmed-nt nb”, “3’D-REGION trimmed-nt nb”, and “5’J-REGION trimmed-nt nb” for a sequence with one D gene and allele. Numbers of mutated nt compared to the germline (underlined in “Analysis of the JUNCTION”, here) are indicated in the dedicated columns of the CSV file: “3’V-REGION mut-nt nb”, “D-REGION mut-nt nb”, “D1-REGION mut-nt nb”, “D2-REGION mut-nt nb”, “D3-REGION mut-nt nb”, “5’J-REGION mut-nt nb” (seq 2: accession number AB012909 of IMGT/LIGM-DB).
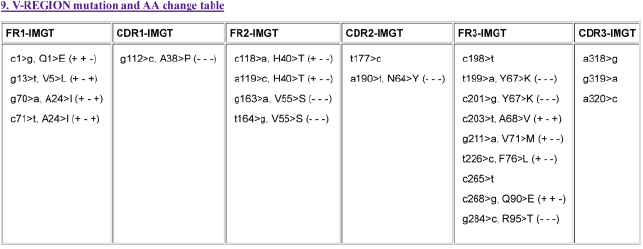
Figure 5. The results of the IMGT/HighV-QUEST “V-REGION-mutation-and-AA-change-table” file #7 (11 columns) are equivalent to those of IMGT/VQUEST “9. V-REGION mutation and AA change table” shown here. As an example, in the FR1-IMGT column, c1>g, Q1>E (++-) means that the nt mutation (c>g) at nt 1 leads to an AA change (Q>E) at codon 1, with same hydropathy (+) and volume (+) classes but different IMGT physicochemical (-) classes [29] (accession number AB012909 of IMGT/LIGM-DB)
6. The “V-REGION-mutation-hotspots” file #10 (8 columns) includes the hot spots motifs ((a/t)a, t(a/t), (a/g)g(c/t)(a/t), (a/t)(a/g)c(c/t)) identified in the closest germline V-REGION and with CDR-IMGT and FR-IMGT localizations. The results of file #10 are equivalent to those of IMGT/V-QUEST “11. V-REGION mutation hot spots” (Figure 7).
7. The “Parameters” file #11 includes the date of the analysis, the IMGT/V-QUEST programme version, the IMGT/V-QUEST reference directory release, and the parameters used for the analysis: the species, the receptor type or locus, the IMGT reference directory set, “with allele *01” (if selected), “Search for insertions and deletions”, and the number of nucleotides to add (or exclude) in 3’ of the V-REGION for the evaluation of the alignment score, and the number of nucleotides to exclude in 5’ of the V-REGION for the evaluation of the number of mutations, if these 2 numbers are not 0 (default value).
IMGT-ONTOLOGY and IMGT® data and information system, which are at the origin of immunoinformatics [45], provide the concepts, knowledge and informatics frame for a standardized analysis of the IG and TR repertoire analysis. IMGT-ONTOLOGY allowed human IG and TR genes and alleles to be identified, described and classified in IMGT® [3-6], before the human genome was published [97,98]. The IMGT Repertoire and the IMGT reference directories are constantly updated with newly sequenced IG and TR of vertebrate species [99-105] and were recently enriched with new human IG genes and alleles owing to the analysis of different haplotypes and CNV in humans [106,107]. These biocurated data and the standardized IMGT® analysis tools are key for maintaining exploration of the immune repertoires with high-quality data at the forefront of basic and clinical research. Indeed, the IMGT® standards for IG and TR are used in clinical applications. IMGT/V-QUEST is frequently used by clinicians in order to identify the repertoires against pathogens and for the analysis of IG somatic hypermutations in leukemia, lymphoma and myeloma, and more particularly in chronic lymphocytic leukemia (CLL) [16,72–75] in which the percentage of mutations of the rearranged IGHV gene in the VH of the leukemic clone has a prognostic value for the patients. For this evaluation, IMGT/V-QUEST is the standard recommended by the European Research Initiative on CLL (ERIC) for comparative analysis between laboratories [72]. The sequences of the V-(D)-J junctions determined by IMGT/Junction Analysis [19,20] are also used in the characterization of stereotypic patterns in CLL [73,74] and for the synthesis of probes specific of the junction for the detection and follow-up of minimal residual diseases (MRD) in leukemias and lymphomas. A new era is opening in hemato-oncology with the use of NGS for analysis of the clonality and MRD identification, and making IMGT®standards use needed more than ever. These standards are also used in the development and description of therapeutical antibodies [40-42,82-86] and the closest human V, J and C genes and alleles (results obtained with the IMGT/DomainGapAlign tool [25-26], approved by the WHO/IUIS nomenclature subcommittee for IG and TR amino acid sequences) are required for the INN submission of humanized and human antibodies [48]. These are the same standards as those utilized by IMGT/HighV-QUEST. The NGS analysis of IG and TR repertoires in physiological conditions [108,109], vaccination [24], autoimmune diseases [110-111] and infectious diseases [112-115] opens new insight for understanding the protective and pathogenic adaptive immune responses. The aims are to characterize the IG and TR repertoires from B and T subsets in many individuals, to identify the skewing of the B and T cell repertoires in immunodeficiency or in autoimmune diseases, to explore the elicitation of broadly neutralizing antibodies (bnAbs) against HIV-1, to understand the potential of the human immune system to develop protective and pathogenic antibodies. The ultimate goal is to find specific antibodies and T cell receptors which could be of great value for diagnostic, prognostic, vaccine development and novel immunotherapy.

Figure 6. The results of the IMGT/HighV-QUEST file #8 (130 columns) and file #9 (109 columns) are equivalent of those of IMGT/V-QUEST “10. V-REGION mutation and AA change statistics” two tables shown here. The results are given for the V-REGION and for FR-IMGT and CDR-IMGT. Statistics are calculated up to the 3’ end of the V-REGION identified in the input sequence (this includes the 3’ last two identical nucleotides with the closest germline V-REGION). The numbers in parentheses, in the V-REGION and CDR3-IMGT columns, correspond to the statistics calculated up to the 3’ end of the closest germline V-REGION and therefore may include nt and AA differences due to the junction diversity (accession number AB012909 of IMGT/LIGM-DB).
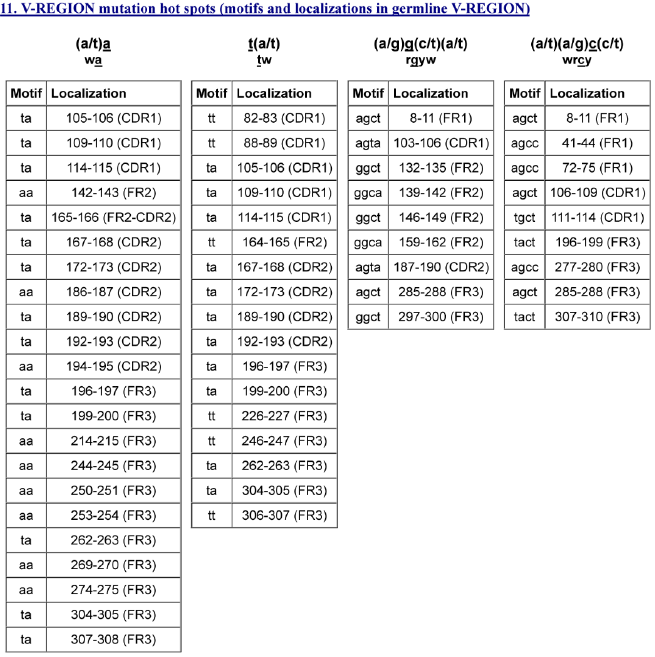
Figure 7. The results of the IMGT/HighV-QUEST “V-REGION-mutation-hotspots” file #10 (8 columns) are equivalent to those of IMGT/V-QUEST “11. V-REGION mutation hot spots” shown here.
We are grateful to Joumana Jabado-Michaloud, Géraldine Folch, Mélanie Arrivet, Pascal Bento, Emilie Carillon, Hugo Duvergey, Amélie Houles, Typhaine Paysan-Lafosse, Marine Peralta, Souphatta Sasorith for their expertise and constant motivation, Gérard Lefranc for his helpful comments, and all the previous members of the IMGT® team, for their invaluable contribution. We thank Cold Spring Harbor Protocol Press for the pdf of the IMGT Booklet available in IMGT references. IMGT® is a registered trademark of CNRS. IMGT® is member of the International Medical Informatics Association (IMIA) and of the Global Alliance for Genomics and Health (GA4GH). IMGT® was funded in part by the BIOMED1 (BIOCT930038), Biotechnology BIOTECH2 (BIO4CT960037), 5th PCRDT Quality of Life and Management of Living Resources (QLG2-2000-01287), and 6th PCRDT Information Science and Technology (ImmunoGrid, FP6 IST-028069) programmes of the European Union (EU). IMGT® is the thematic ELIXIR Immunoinformatics node of the Institut Français de Bioinformatique IFB. IMGT® is currently supported by the Centre National de la Recherche Scientifique (CNRS), the Ministère de l’Enseignement Supérieur et de la Recherche (MESR), the Montpellier University, the Agence Nationale de la Recherche (ANR) Labex MabImprove (ANR-10-LABX-53-01), BioCampus Montpellier, the Région Languedoc-Roussillon (Grand Plateau Technique pour la Recherche (GPTR). This work was granted access to HPC@LR and to the High Performance Computing (HPC) resources of Centre Informatique National de l’Enseignement Supérieur (CINES) and to Très Grand Centre de Calcul (TGCC) of the Commissariat à l’Energie Atomique et aux Energies Alternatives (CEA) under the allocation 036029 (2010-2015) made by GENCI (Grand Equipement National de Calcul Intensif).
Authors who use IMGT® databases and tools are encouraged to cite this article and to quote the IMGT® Home page, http://www.imgt.org. Online access to IMGT® databases and tools are freely available for academics and under licenses and contracts for companies.
The authors declare no conflict of interest.
Download Provisional PDF Here
Article Type: Research Article
Citation: Giudicelli V, Duroux P, Lavoie A, Aouinti S, Lefranc M-P, et al. (2015) From IMGT-ONTOLOGY to IMGT/HighV-QUEST for NGS Immunoglobulin (IG) and T cell Receptor (TR) Repertoires in Autoimmune and Infectious Diseases. Autoimmun Infec Dis 1(1): doi http:// dx.doi.org/10.16966/2470-1025.103
Copyright: © 2015 Giudicelli V, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access