Abstract
Background: Our group has used deep sequencing to identify viral RNA signatures in human brain specimens. We have previously used this method to detect HSV1, GBV-C, and measles virus sequence in brain tissue from deceased donors. Deep sequencing was performed on brain specimens from a cohort of patients who died with progressive forms of MS, revealing evidence of increased expression of some human endogenous retrovirus (HERV) domains.
Objectives: Identify RNA sequences and new antigens involved in the pathogenesis of MS
Methods: Deep sequencing was performed on RNA extracted from 12 progressive MS, 2 neuromyelitis optica (MS/NMO = demyelination group), 14 normal control, and 7 other neurologic disease (OND) control frozen brain specimens. The resulting single-ended 50 bp sequences (reads) were compared to a non redundant viral database representing (NRVDB) all 1.2 M viral records in GenBank. A retroviral gene catalog (RVGC) was prepared by identifying human genetic loci (GRCh37.p13) homologous to domains contained in the Gypsy 2.0 retro element database. Reads were aligned to the RVGC and human transcriptome with Bowtie2. The resulting viral hit rates (VHRs) were normalized by the number of high quality reads. The expression of human genes, including HERVs, was determined using Cufflinks. Comparisons between the groups were performed using the false discovery rate.
Results: Fifty to 131 million high quality reads per specimen were obtained. Comparison of the reads to the NRVDB suggested that the demyelination and OND specimens had higher VHRs against some retroviral sequences compared with the controls. This was confirmed by retroviral domain averaging. Gene expression analysis showed differential expression among some HERV sequences. Single read mapping revealed one envelope and one reverse transcriptase sequence record that were significantly enriched among the demyelination samples compared to the normal controls. Less restrictive (comprehensive) read mapping showed that 2 integrase, 2 core, 2 envelope, and 3 KRAB sequences that were overexpressed in the demyelination group.
Conclusions: These data demonstrate that some endogenous retroviral sequences are significantly overexpressed in these demyelination brain tissue specimens, but the magnitude of this overexpression is small. This is consistent with the concept of HERV activation as a part of the innate immune response.
Keywords
Deep sequencing; RNA-seq; Next generation sequencing; Endogenous retroviruses; HERV; Multiple sclerosis; Progressive MS
Introduction
Multiple sclerosis (MS) is a chronic demyelinating disease of unknown cause, which affects the brain and spinal cord of about 400,000 individuals in the U.S. A number of viral infections of the CNS can lead to demyelination, including distemper (dogs), measles (subacute sclerosing panencephalitis, SSPE, humans), and influenza (humans). [1]. Viruses have long been suspected as causative agents of MS, based on the epidemiology of the disease including geographic patterns, isolated outbreaks, and migration studies [2-5]. Novel viruses that cause human disease continue to be discovered including hepatitis C (1989), corona virus NL63 (2004), bocavirus (2005), and rhinovirus C group (2007) [6- 9]. Novel human polyoma and arena viruses were recently identified as causes of serious human diseases by deep sequencing [10-12]. The National Multiple Sclerosis Society itself provides an excellent rationale for the search for viruses in MS and related diseases [13].
Past serology studies provided some evidence for the involvement of retroviruses in the pathogenesis of MS [14,15]. Other groups have identified human endogenous retroviruses (HERVs), including HERV Fc1 and HERV-W, as possible MS pathogens [16-24]. Polymorphisms of human genes involved in the control of retroviral replication, including TRIM5, TRIM22, and BST2, are associated with higher risk of developing MS [25] and recent epidemiologic studies suggest that MS and HIV are mutually restrictive; that is, HIV patients develop MS less frequently than expected, adjusted for age, sex, and other factors [26-28].
Our group has used deep sequencing (also called next generation sequencing) to detect microbial sequences in donor brain tissues. This powerful new technique allowed identification of GBV-C in the brain of one MS subject and HSV or measles virus in the brains of several persons with encephalitis [29,30]. Deep sequencing is applied here to cryopreserved progressive MS and NMO brain specimens in comparison to normal brain and encephalitis brain specimen controls.
Methods
Primary progressive MS cases were requested from the Human Brain and Spinal Fluid Resource Center (Los Angeles Veterans Administration, California) and the Rocky Mountain MS Center (Englewood, Colorado) brain repositories. Cases were selected by the directors of these two institutions. Specimens were studied from 11 persons with primary progressive MS (PPMS), one with secondary progressive MS (SPMS), and 2 persons with severe progressive neuromyelitis optica (NMO). Progressive MS and NMO cases were specifically selected because these tend to be the most severe subtypes of the MS-related diseases. Fourteen normal control and 5 frozen encephalitis brain specimens these resources were also studied. Two additional de-identified frozen encephalitis specimens were obtained from Dr. Don Gilden at the University of Colorado, Denver. The specimens were collected post-mortem within one day of death, either fresh frozen or snap frozen in liquid nitrogen, and were associated with a neuropathologic diagnosis. The research plan was submitted to the University of Utah Health Sciences IRB for review. This research was reviewed and approved by the University of Utah Health Sciences IRB, #00028658.Since this research involved only de-identified post-mortem material, it was found to be exempt from consent, review, and oversight.
The samples were assigned to one of three groups:
- Demyelination (N=14)
- Primary progressive MS (N = 11)
- Secondary progressive MS (N = 1)
- Neuromyelitis optica (N = 2)
- Normal controls (n=14)
- Other Neurologic Disease (OND) (n=7)
- Herpes encephalitis (N = 3)
- Other/unknown encephalitis (N = 2)
- Subacute sclerosing pan encephalitis (N = 2)
The frozen brain specimens were handled as previously described [29] RNA was extracted from frozen brain with the RNeasy Lipid Tissue Mini Kit (Qiagen, Valencia, CA, Cat No./ID: 74804). The extracted RNA was DNase treated by incubating 15 minutes on the column. The resulting RNA was submitted for sequencing at the University of Utah High Throughput Genomics Shared Resource. Prior to sequencing, RNA was analyzed on an Agilent Bioanalyzer Nano chip (Agilent Technologies, USA) and evaluated for RNA abundance and integrity as previously described [29] Samples were reverse transcribed and libraries were prepared with the Illumina TruSeq kit (Illumina, San Diego, CA). To ensure the inclusion of possible viral RNA genomes, oligo dT selection was not performed. To avoid possible enrichment intrinsic biases, rRNA depletion was not performed.
The samples were sequenced and the sequencing reads were processed and aligned as previously described [29] Briefly, the reads were Illumina HiSeq 2500, 50 bp single-end. FASTQ format sequences were qualityfiltered and high quality (HQ) sequences were retained. Reads that aligned to either the human genome (NCBI build GRCh37.p13) or human transcriptome, were removed from the HQ reads, yielding “screened reads” [31] Screened reads were aligned to the non-redundant viral database (NRVDB) using MegaBLAST (v2.2.26) with a word size of 28 bp [32] Viral hit counts (reads aligned to sequences in NRVDB) were normalized (divided) by the number of (HQ) high quality reads in a specimen.
The retroviral gene catalog (RVGC) was prepared by identifying regions of the human genome (build GRCh37.p13) with detectable similarity to core retroviral domains in the Gypsy 2.0 database (GyDB) using BLASTX [33,34] Alternative retroviral databases were considered for this analysis, and Gypsy was chosen due to the presence of annotations that allowed categorization of the sequences into retroviral domains. The BLASTx subprogram of version 2.2.26 of NCBI blast all was used with default word size and scoring and an expect cutoff of 0.1. Human genomic alignments with length 50% or longer than the subject retroviral domain were placed in RVGC. RVCG entries were named with arbitrary unique codes of the form: _U (see S1 RVGC Table). For record keeping, the GyDB domain that was aligned to human sequence derived RVGC entry (recognition domain) and the source GI, starting position and length are also encoded in each RVGC fast a record name. Specifics of the compiled RVGC are available in Supplementary Data. Expression normalization was determined using the expression analysis pipeline Bowtie-Tophat-Cufflinks using only annotated splice sites [35]. The cufflinks norm mass parameter was used as the number of fragments for all FPKM metrics.
Statistical analysis of the demographic characteristics of the study population was performed using Vassar Stats [36], an open-source web application, and Microsoft Excel for Mac 2011. Differences among the groups were screened for significance by one way un-weighted ANOVA testing for continuous variables (age, year of collection, and post-mortem interval), and the two-tailed Chi-squared test with Yates correction for discrete variables (sex). For each taxon, viral hit rates (VHR) were compared between each demyelination and OND specimen and the set of controls using the Z-test with Bonferroni correction for multiple comparisons. Taxa where none of the demyelination or OND specimens VHRs were significantly different from controls were excluded from further analysis. Specific differences in VHRs between the groups were tested using the Mann-Whitney U-test or Tukeys HSD test. Correction for multiple comparisons was accomplished using the Benjamini– Hochberg procedure with a false discovery rate (FDR or q) of 0.05 [37]. The distribution of the sample types in the Retroviral Domain Discrimination Analysis was tested using the two-tailed Fishers Exact test for 2 × 2 tables [36].
Results
Subject Demographics
Characteristics of the study samples are shown in table 1. Fourteen demyelination samples were compared to 14 normal controls and 7 OND specimens. All patients in the demyelination group had severe and progressive clinical disease. Eleven were categorized as PPMS, two as NMO, and one as secondary progressive MS. The postmortem interval (PMI), time between death and collection of the brain sample, range was 2-26 hours. There were no significant differences in PMI between the groups (p=0.66). Age and sex information was available for all 14 controls, 13 of 14 demyelination cases, and 5 of 7 OND cases. The proportion of known females in the demyelination group 9/13 (69%) was not significantly different than the control group 6/14 (43%) (p=0.32). Ages of the subjects ranged from 37-93 years. The demyelination group was significantly younger (mean age 58 ± 13 years) than the normal controls (mean 71 ± 12 years, p=0.017). The specimens were collected between 1981 and 2010. The year of collection was not significantly different between the groups (p=0.29).
| Group |
Specimen |
PMI |
Age |
Sex |
Collection Year |
Clinical History |
Specimen Location |
Neuropathology Reading |
HQ
Reads |
| Normal Control |
202 |
4 |
57 |
M |
1983 |
Post-surgical infection |
cerebrum |
normal |
91.5 |
| 214 |
4 |
56 |
M |
1981 |
Heart Disease |
cerebrum |
normal |
94.7 |
| 3276 |
19 |
54 |
M |
2002 |
Heart Disease |
frontal WM |
normal |
93.9 |
| 33 |
5 |
69 |
M |
1983 |
Lung Disease |
cerebrum |
normal |
89.1 |
| 3371 |
16 |
52 |
M |
2002 |
Lung Cancer |
frontal WM |
normal |
96.8 |
| 3406 |
20 |
72 |
F |
2002 |
Heart Failure |
temporal WM |
normal |
89.3 |
| 3465 |
20 |
93 |
F |
2002 |
Bleeding |
temporal WM |
normal |
105.2 |
| 3482 |
14 |
79 |
F |
2003 |
Heart Disease |
temporal WM |
normal |
97.5 |
| 3543 |
12 |
73 |
F |
2003 |
Lung Disease |
parietal WM |
normal |
105.2 |
| 3348 |
9 |
76 |
F |
2002 |
Heart Disease, Diabetes |
frontal WM |
normal |
102.7 |
| 3465 |
11 |
76 |
M |
2003 |
Heart Failure |
occipital WM |
normal |
107.3 |
| 3637 |
13 |
76 |
M |
2003 |
Pneumonia |
frontal WM |
normal |
102.5 |
| 3641 |
20 |
76 |
M |
2003 |
Stomach and Liver Cancer |
parietal WM |
normal |
85.3 |
| 3698 |
17 |
84 |
F |
2003 |
Breast Cancer |
temporal WM |
normal |
90.3 |
|
Demyelination (Progressive MS and NMO) |
168 |
3 |
37 |
F |
1984 |
PPMS |
cerebrum |
reactivated MS |
89.7 |
| 2443 |
26 |
49 |
F |
1997 |
PPMS |
periventricular WM |
lymphocytic cuffing |
99 |
| 2485 |
9 |
69 |
M |
1997 |
PPMS |
frontal WM |
active MS |
63.3 |
| 2696 |
21 |
86 |
F |
1998 |
PPMS |
periventricular WM |
chronic MS plaque |
130.9 |
| 2946 |
15 |
59 |
M |
1999 |
PPMS |
periventricular WM |
chronic perivascular
inflammation |
88.9 |
| 3161 |
20 |
51 |
F |
2001 |
Secondary Progressive MS |
periventricular WM |
active MS with
lymphocytic cuffing |
89.9 |
| 3185 |
14 |
50 |
M |
2001 |
PPMS |
periventricular WM |
chronic MSplaque |
91.1 |
| 3509 |
11 |
74 |
F |
2003 |
PPMS |
periventricular WM |
chronic MS plaque |
87.2 |
| 3816 |
21 |
47 |
F |
2004 |
PPMS |
frontal WM |
macrophages and
lymphocytic cuffing |
91 |
| 3840 |
23 |
61 |
F |
2003 |
PPMS |
frontal WM |
macrophages and
lymphocytic cuffing |
95 |
| 3931 |
10 |
74 |
F |
2004 |
PPMS |
periventricular WM |
chronic active disease |
73.8 |
| 5149 |
8 |
48 |
M |
2010 |
PPMS |
frontal WM |
active MS |
66.4 |
| 108 |
2 |
56 |
F |
1998 |
NMO |
midbrain and pons |
demyelination with necrosis |
72.5 |
| 290 |
NA |
NA |
NA |
NA |
NMO |
corpus callosum |
demyelination consistent with NMO |
82.3 |
| OND Control |
1418 |
5 |
68 |
M |
1988 |
MS-like illness† |
Frontal cortex |
chronic encephalitis |
76.8 |
| 4403* |
18 |
77 |
F |
2006 |
HSV encephalitis, stroke |
Frontal cortex |
active encephalitis |
99.5 |
| 4471 |
12 |
73 |
F |
2007 |
Rasmussen’s encephalitis |
Frontal cortex |
gliosis without active
inflammation |
49.4 |
| 710* |
8 |
58 |
M |
1983 |
HSV encephalitis, chronic lymphocytic leukemia |
Frontal cortex |
active encephalitis |
50.1 |
| 924* |
24 |
86 |
M |
1985 |
HSV encephalitis |
Frontal cortex |
chronic encephalitis |
67.9 |
| coloradoA |
NA |
NA |
NA |
NA |
SSPE (measles) |
NA |
NA |
67.9 |
| coloradoB |
NA |
NA |
NA |
NA |
SSPE (measles) |
NA |
NA |
66.9 |
NA = information not available
NMO = neuromyelitis optica
OND = other neurologic disease
PMI = post-mortem interval (hours)
HQ reads = high quality reads (in millions)
PPMS = primary progressive multiple sclerosis
SSPE = subacute sclerosing panencephalitis (measles infection)
WM = white matter
*previously diagnosed as HSV encephalitis
†MS not confirmed by pathology
Table 1: Characteristics of the sequenced frozen brain specimens.
Sequencing and Analysis
Deep sequencing yielded between 49.4 and 130.1 million 50 bp high quality (HQ) sequences per sample. The mean yield of HQ sequences was not different between the normal control (µ = 96.5 ± 7.1M) and demyelination (µ = 87.2 ± 16.6M) groups. The OND group yielded significantly fewer HQ sequences per sample (µ = 68.3 ± 17.0M) than both the normal control and the demyelination groups (p<0.01 for both comparisons).
A heat map showing log-transformed HRs was generated from the sequencing analysis (Figure 1) [38]. Only viral taxa where one or more specimens are significantly overrepresented are displayed in the figure. Fifty viral taxa were over represented in at least one of the demyelination or OND specimens compared to the set of normal controls. In this manner, false positive alignments to human and cloning sequence were removed. GB Virus C, previously shown to be present only in specimen 3840, was the only non-retrovirus definitively present in any of the demyelination samples [30]. Bona fide and spurious viral alignments within the OND samples have been previously described [29].
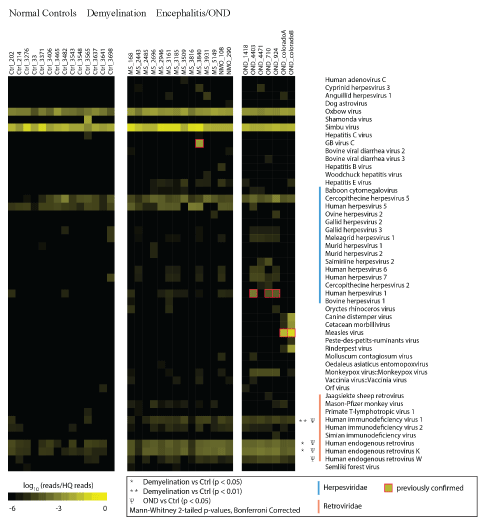
Figure 1: Hit Rate Comparison of Frozen Brain Specimens.
RNA was extracted from 14 normal control, 14 demyelination (including 2 NMO), and 7 other neurologic disease (OND) cryopreserved brain specimens. cDNA libraries were prepared and deep sequencing was performed. High quality sequences were matched to the non-redundant viral database using Mega BLAST with a word size of 28 bp. Hits (matches to the database) were normalized for the total number of high quality reads in each specimen and log10 transformed, providing a ‘Log Hit Rate.’ Sequences mapping to non-vertebrate viruses (phages, plant and insect viruses) and cloning vectors were removed. Viral taxa not significantly enriched in any of the samples (e.g. VZV, EBV, and Enteroviruses) are not shown.
Among the 50 viral taxa overrepresented in at least one OND or demyelination samples, 17 were herpes viruses and 9 were retroviruses. Statistical comparisons (corrected for multiple comparisons) of VHRs between all members of the groups were performed. This analysis revealed that only 3 retroviral taxa that were significantly over expressed in the demyelination group compared to the control group: human immunodeficiency virus 1 (HIV-1), human endogenous retroviruses (family), and human endogenous retrovirus K. There were no AIDS or HIV patients in the cohort; thus the high VHR to HIV-1 was inferred to be caused by the 50 bp reads aligning to sequences similar to HIV-1. None of the 9 herpes virus candidate taxa were significantly different between the groups.
Retroviral Analysis
The apparent retroviral sequence enrichment in the demyelination group led to a more inclusive retroviral sequence analysis. A new database called the retroviral gene catalog (RVGC) was prepared (see Methods). RVGC contains endogenous sequences similar to protein-coding retroviral genes: GAG, RT, ENV, etc. Specific information about all the sequences records in the RVGC, including GenBank identifiers, length, and location is available (S1 RVGC Table).
Reads aligning to the RVGC database were binned according to retroviral domains type (e.g. GAG, RT, ENV). GAG refers to the viral core, RT to the reverse transcriptase, and ENV the envelope. The functions of the SCAN domain are not well understood and KRAB is probably a transcriptional repressor [39]. CHR refers to the chromo domain found at the C-terminal end of many retro transposon integrases [40].
Mean domain-type hit rates (HRs) for the demyelination, NMO and control samples were log2 transformed and centered in the domaintype axis (Figure 2). These values were hierarchically clustered (Pearson correlation) and compared between the demyelination and normal control specimens [41]. The resulting sample clustering reveals the relative separation of demyelination and control specimens into the dominant nodes of the cluster (e.g. 9/10 demyelination samples cluster left, 12/16 control samples cluster right; p=0.004). This data set shows evidence of broad retroviral gene over expression in the brains of the demyelination subjects compared with normal controls. However, the magnitude of the retroviral over expression was small, less than 2-fold, and was not evident for any single RVGC sequence. This retroviral gene expression pattern was more pronounced among the OND (encephalitis) samples than in the demyelination group. Expression of the neural tissue control genes RPL13, RPL19, and UBC was not significantly different between the 3 groups.
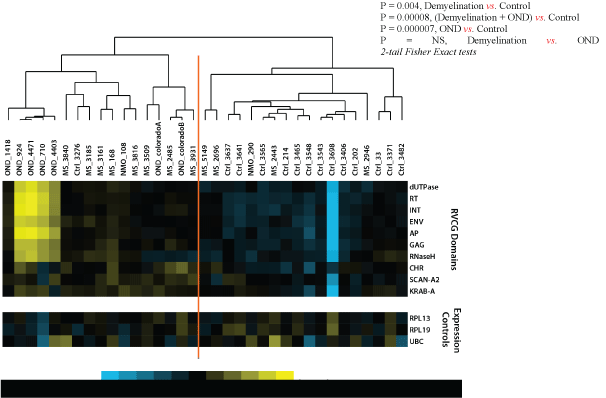
Figure 2: Retroviral Domain Discrimination Analysis.
Sequencing hits (matches) to the RVGC database were binned into retroviral domains. Mean log2 transformed and centered hit rates to the retroviral domain types in RVGC for the demyelination (MS), OND and normal control specimens. The results are clustered using Cluster 3.0 [41], showing distinctly different expression patterns in most demyelination, control and OND specimens.
Additional analysis showed that some specific retroviral genes are significantly over expressed in the demyelination (N=14) and PPMS only (N=11) groups compared with the normal controls (n=14). Two mapping procedures were employed: “Best Alignment” where each read was mapped to the RVGC only once to its best match, and “Comprehensive Alignment” where every reported Bowtie 2 alignment was counted. The results of this analysis are displayed in Table 2. Only one envelope gene and one RT gene were significantly over expressed by the best alignment procedure. Several integrase, GAG, and envelope genes, along with 3 KRAB genes, were over expressed by the comprehensive alignment procedure; limiting the analysis to the 11 PPMS specimens showed those 2 ENVs and 3 GAGs were over expressed compared to controls. The HERV annotations for these over expressed genes are shown in Table 2.
| Method |
Group |
Gene |
Expression 1 |
Ratio2 |
P-value |
FDR (q) D:C3 |
Recognition Domain |
|
Best Alignment |
Demyelination |
ENV-U3 |
958 |
1.7 |
0.0001 |
0.006 |
K-HERV |
| (N=14) |
RT-U105 |
300 |
1.7 |
0.0001 |
0.032 |
MMTV |
| PPMS only |
ENV-U3 |
952 |
1.7 |
0.0002 |
0.017 |
K-HERV |
| (N=11) |
RT-U105 |
311 |
1.8 |
0.00005 |
0.014 |
MMTV |
|
Comprehensive Alignment |
Demyelination |
INT-U176 |
775 |
1.7 |
0.0001 |
0.036 |
MuERV-L |
| (N=14) |
INT-U45 |
94 |
2.8 |
0.0003 |
0.044 |
|
| |
GAG-U21 |
52 |
2.5 |
0.0003 |
0.009 |
K-HERV |
| |
GAG-U22 |
43 |
2.3 |
0.0011 |
0.018 |
HERV-K10 |
| |
ENV-U59 |
1540 |
1.9 |
0.0009 |
0.037 |
RTVL-Ia |
| |
ENV-U3 |
960 |
1.7 |
0.00007 |
0.006 |
K-HERV |
| |
KRAB-U15 |
9880 |
1.8 |
0.001 |
0.017 |
KRAB |
| |
KRAB-U13 |
1027 |
1.7 |
0.001 |
0.017 |
|
| |
KRAB-U9 |
1982 |
1.5 |
0.006 |
0.037 |
|
| PPMS only |
ENV-U3 |
952 |
1.7 |
0.0002 |
0.019 |
K-HERV |
| (N=11) |
ENV-U59 |
1617 |
2 |
0.0009 |
0.04 |
RTVL-Ia |
| |
GAG-U21 |
56 |
2.7 |
0.00009 |
0.003 |
K-HERV |
| |
GAG-U22 |
47 |
2.5 |
0.0006 |
0.009 |
HERV-K10 |
| |
GAG-U8 |
242 |
1.5 |
0.004 |
0.041 |
HERV-E |
1Expression is described as FPKM (Fragments Per Kilobase of transcript per Million mapped reads) × 1000
2Ratio = mean expression in the Demyelination or PPMS group divided by mean expression in the Control group
3False Discovery Rate (FDR) q-values were calculated over each domain type separately (e.g. RT, ENV, GAG, etc.)
Table 2: Specific Retroviral Genes Overexpressed in the Demyelination Group: Fifty bp reads were aligned to all annotated HERV sequence in NCBI/GenBank using the Bowtie2application with the default settings for end to end alignment. Mean expression levels (reads per million per kilobase,
FPKM) are shown for the demyelination (progressive MS and NMO) groups. Reads were mapped either: 1) singly, to their best matching RVCG sequence
(Best Alignment) or, 2) using every reported Bowtie2 alignment to the RVGC (Comprehensive Alignment). P-values and False Discovery Rates (FDR) are
reported, along with the ratio of the mean hit rates between the experimental and control groups. Retroviral associations are per the annotation in the
Gypsy 2.0 database.
Discussion and Conclusion
This study employed deep sequencing and metagenomic analysis techniques to comprehensively investigate retroviral expression among frozen demyelination brain samples compared with normal controls and OND (encephalitis) controls. The results show some over expression of HERVs in general among most domains (Figure 2). Over expressed HERV and KRAB sequences were specifically identified corresponding to several retroviral domains, including core, envelope, integrase, and reverse transcriptase (Table 2). These results support the hypothesis that retroviral sequences are over expressed in demyelinated brain samples compared with normal brain. However, the magnitude of the observed retroviral domain over expression was small, less than 3-fold, and the pathological significance of this observation is unknown.
The data from this study are consistent with the concept that HERV over expression is part of the human immune response. Other groups have identified the MSRV (HERV-W) or HERV-Fc1 as possibly contributing to the pathogenesis of MS [16,17,19,20,24,42]. Interestingly, the present sequencing study did not specifically confirm these findings (Figure 1). Instead, some other HERV genes from a variety of sources were shown to be significantly over expressed (Table 2). This highlights the difficulties inherent with these studies where multiple similar HERVs have been incorporated into the human genome. The gene expression mapping performed here relies on annotated sequences from the human genome build 37. The data generated in the present sequencing study is comprehensive across the entire human genome, but it is likely to be less specific and less quantitative than qPCR.
Human endogenous retroviruses (HERVs) are remnants of ancient retroviral infections of the host germ line that are transmitted vertically from parents to their offspring. The utility of these elements within the human genome remains largely speculative, although at least one HERV codes for syncitin, an important protein that allows for development of the placenta [43].
Interestingly, some animal ERVs efficiently interfere with the replication of related exogenous retroviruses [44,45]. Sheep have been used to study the evolution of ERVs within a mammalian host due to the presence of related exogenous and endogenous retroviruses. The exogenous (i.e., horizontally transmitted) oncogenic retrovirus, Jaagsiekte sheep retrovirus (JSRV), causes fatal lung cancers. A closely related provirus, JSRV-20, entered the host genome within the last 3 million years during speciation within the genus Ovis. Endogenous JSRV has a defective Gag polyprotein resulting in a transdominant phenotype that blocks the replication of the closely related exogenous JSRV [46-48]. That is, the expression of an endogenous retrovirus effectively blocks a fatal infection with an exogenous retrovirus. This animal data strongly suggests that endogenization and selection of ERVs is a mechanism used by the host to fight retroviral infections. Support for this concept in humans is displayed by the increased expression of (endogenous) HERV-K in patients infected with the (exogenous) retrovirus HIV, where HERV-K envelope is neuroprotective [49,50].
Most of OND specimens were from patients with either herpes (3) or measles virus (2) encephalitis. The HSV infected specimens (4403, 710, 924) displayed the highest levels of HERV domain expression, as shown in Figure 2. This is consistent with the results of other groups that have shown HSV stimulates reverse transcriptase in PBMCs from MS patients [51], and HERV-W expression is induced by HSV1 in cell cultures [52,53].
One limitation of this deep sequencing method is that the RNA extractions are not completely DNA free, despite a DNAse treatment step. Qubit analysis of extracted brain specimens revealed that 1-5% of the analyzed material is DNA retained from the original sample. Prior to sequencing, the RNA were also analyzed on an Agilent Bioanalyzer Nanochip (Agilent Technologies, USA) and evaluated for RNA size, abundance and integrity. This provided relatively high quality RNA for the subsequent analysis, but it cannot be determined with absolute certainty that retained DNA did not affect the results of the study. Another limitation of the study is the post-mortem interval necessarily associated with these samples obtained from deceased human donors. While there was no detectable difference in the PMI between the demyelination and control specimens, this interval likely allowed some RNA to degrade in all the samples. This likely did interfere with the detection of HERV and other retroviral sequences during the sequencing reactions.
Acknowledgments
The authors would like to acknowledge help from Dr. Rashed M. Nagra, Director, Human Brain and Spinal Fluid Resource Center (UCLA Brain Bank), Los Angeles, CA; Dr. John Corboy, Director, Rocky Mountain MS Center, Denver, CO; and Dr. Don Gilden at the University of Colorado for the provision of frozen brain specimens. Brian Dalley from Huntsman Cancer Institute Microarray Core Facility supervised the preparation of cDNA libraries and the Illumina sequencing.
Funding
This work was funded by grant R21NS077023 from the NIH/NINDS, “Deep Sequencing for the Detection of Viral Sequences in Primary Progressive Multiple Sclerosis Brains.” Some follow-up work was funded by the National MS Society, grant #RG4807A4.