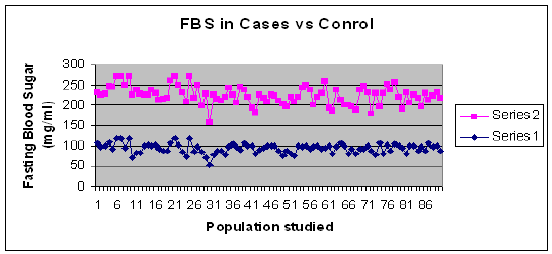
Graph 1: Comparison of FBS levels in T2DM Cases and Control

Alice JayaPradhaCheekurhty*1 C Rambabu2 Amit Kumar3
1Research Scholar, Department of Biochemistry, Acharya Nagarjuna University, Nagarjuna Nagar, Guntur, Andhra Pradesh, India*Corresponding author: Alice Jaya Pradha Cheekurhty, Research Scholar, Department of Biochemistry, Acharya Nagarjuna University, Nagarjuna Nagar, Guntur, Andhra Pradesh, India, Tel: 9581771118; E-mail: alicejaya@gmail.com
The aim of this study was to analyze the proven role of candidate genes, Calpain10 and Adiponectin allelic variants in genetic susceptibility to type 2 diabetes mellitus in our discrete diabetic population of Andhra Pradesh and Telangana states of India.
In accomplishment of our objective to find the presence of previously reported SNPs we could find not only the consistency of the reported SNPs we also found the new Single Nucleotide Polymorphisms in the Calpain 10 gene.
Type 2 Diabetes Mellitus (T2DM); Calpain 10 (CAPN 10); Adiponectin (ADIPOQ); Single nucleotide polymorphism; Risk Factors
Development of Type 2 Diabetes Mellitus (T2DM) and advancement in form of related complications are due to interplay of Several Genetic, Environmental and Biochemical risk factors [1]. The prevalence of diabetes is increasing at an alarming rate for all age-groups worldwide with a significant effect on adult population [2,3]. The largest increase of 80% (The International Diabetes Federation) is observed in India with people living in southern states more prone due to their life style Characteristic “Asian Indian Phenotype” makes them more susceptible to the disease [4,5]. Several Biochemical [6] and genetic factors [7] are responsible for the outcome of type 2 diabetes mellitus.
This study is a part of case control study for identifying genetic and non-genetic risk factors for T2DM and its related complications. SNPs of seven genes were studied along with the biochemical parameters to find the individual and combined effect of these two risk factors.
The previously reported single nucleotide polymorphisms (SNPs) in Calpain-10 [8-12] (CAPN10) and Adiponectin [13-17] were selected by studying the related work at that time of initiation of our studies.
Calpain10 gene [18] encoded by cysteine protease spans 66 kb on chromosome 2q37.3 with 15 exons and. Its role is in the regulation of a variety of cellular functions and may be responsible for adipocyte differentiation. Its role in glucose regulation is known. This is the first but not the last candidate gene with susceptibility for Type 2 Diabetes Mellitus identified through a positional cloning and genome wide screen [19]. The role of CAPN10 has been examined among different ethnic groups in wide range of population studies with uneven results [20-23]. This is due to genetic heterogeneity between populations. Some confirm the original finding that haplotype combination comprising three intronic CAPN10 single-nucleotide polymorphisms (UCSNP-43, -19, and -63) was associated with increased risk of T2DM but some do not.
Adiponectin belongs to the family of adipokines. It is the product of ADIPOQ gene It is located on Chromosome 3q27. It is the major adipocyte secretory protein most abundantly found in human plasma with potent roles in fat metabolism and glucose regulation. It includes insulin sensitivity in muscle and liver, regulating energy homeostasis and glucose tolerance. The low circulating levels of adiponectin are associated with obesity and diabetes.
The left over peripheral blood sample of subjects tested for routine blood examinations was collected from the diagnostic Centre. They were either self-motivated or prescribed by the doctor. Each of the subjects was subjected to simple questionnaire primarily to have a case history. The samples included were of diabetic and normal healthy subjects aged between 15-85 years from Andhra Pradesh and Telangana States in India. The case subjects of the study were established diabetic patients based on their biochemical reports which showed Fasting Blood Sugar (FBS) value of more than 126 mg/dl or ≥ 7.0 mmol/L and Post Prandial Blood Sugar (PPBS) value of more than 200 mg/dl or ≥ 11.1mmol/L (WHO diagnostic criteria for diabetes).The healthy subjects with FBS and PPBS value less than above values and with no family history of diabetes served as controls.
The peripheral blood samples were collected in sitting position in the EDTA and non-EDTA vials in accordance to the protocol from 200 subjects. 20 subjects suffering from other infectious diseases were excluded from the study and those undergoing medical treatment for T2DM were included in the study. Finally a total of 180 subjects 96 were females and 84 were males were included in the study.
We have investigated the polymorphisms rs2975760 [26] and rs3792267 [27] in CAPN10 gene and rs3774261 [28] in ADIPOQ gene reported for contribution to risk of T2DM in were selected for their validation in our study population.
Out of 180 left over blood samples selected 90 were diabetes cases and 90 were controls. They were analyzed for different biochemical parameters of CBP and lipid profile and of which 51 samples were processed for extraction and purification by using Sambrook et al. protocol [29-31]. The leucocytes component was separated from other cell components by washing in Tris wash buffer, prior to lysis using lysis buffers and proteinase K. All the contaminating red blood cells and the serum proteins are removed in the lysis step. These lysed proteins are precipitated using sodium acetate. The CAPN 10 and ADIPOQ genes with specified target region of SNP associated with T2DM was amplified using Polymerase chain reaction [31,32] from this isolated and purified DNA. The reaction mixture of PCR consists of TaqDNA polymerase enzyme, primers (Forward and reverse), nucleotides and reaction buffer. Primer designing was done on Primer Blast tool. The conditions for PCR were optimized to produce high yields of specific target sequences. The PCR amplified products were qualitatively checked on 1.2% agarose gel electrophoresis [33] and were visualized in transilluminator after staining with ethidium bromide. These amplicons were then subjected to sequencing by Sanger sequencing method [34]. The sequenced portions of the genes were checked for the consistency of reported mutations in our samples.
The sequencing of amplified portion of the two genes revealed the presence of SNPs in some the diabetic population studied by us. We observed that in CAPN10 gene the SNPs rs2975760 and rs3792267 are present in 29 cases. The SNP rs2975760 is present 20 T2DM cases. The nucleotide and corresponding amino acid change C→T (Ala→Val), is noticed in 20 T2DM cases. Similarly the change G→C (Val→Ile) is noticed in rs3792267 (SNP) in another 20 cases.
The nucleotide change and corresponding amino acid change in the ADIPOQ gene is A→G (Met→Ile) for SNP rs3774261 is noticed in 17 T2DM cases. The nucleotide and the corresponding amino acid change A→G (Arg→Gly) in a new position in 13 cases of T2DM is identified. CAPN10 gene SNPs are not present in controls. All the three SNPs are present in 7 of the T2DM cases (Table 1).

Graph 1: Comparison of FBS levels in T2DM Cases and Control
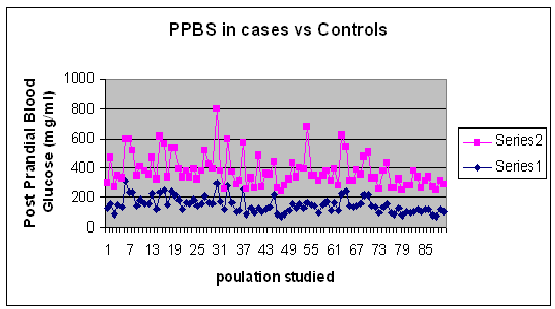
Graph 2: Comparison of PPBS levels in T2DM Cases and Control
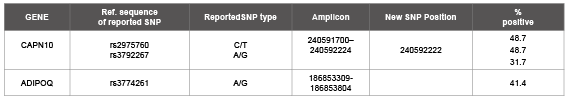
Table 1: Position of Single Nucleotide Polymorphism
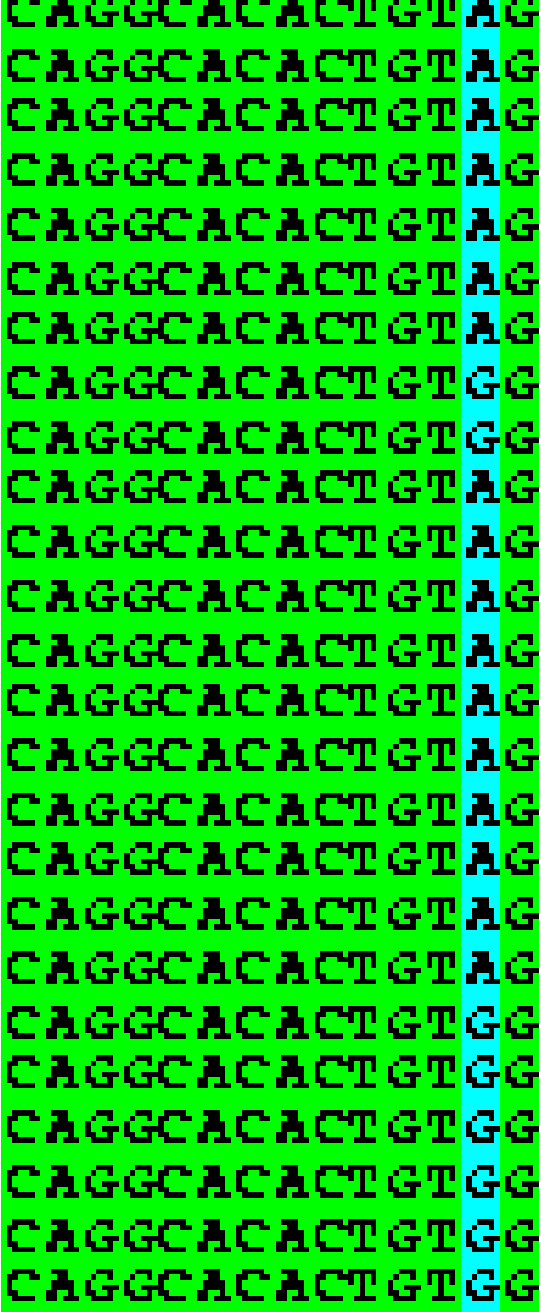
Figure 1: Box Shade of novel SNPs in CAPN10
This case control observational study showed increased Fasting Blood Sugar value of more than 126 mg/dl and Post Prandial Blood Sugar value of more than 200 mg/dl (WHO criteria or diabetes) was observed in Type 2 Diabetes Mellitus cases when compared to the controls.
The mean values for distribution of blood sugar levels is found to be FBS 152 ± 92.8 92 ± 11.7 and p = 0.8 and PPBS 229.3 ± 67.6 131.2 ± 18.8 p = 0.7.The comparison of Fasting Blood Sugar and Post Prandial Blood Sugar is shown as graphical representation below in Graph1 and Graph 2.
Correlation studies showed that most of the subjects with increased blood sugar levels fasting and post prandial levels of glucose were seen in case subjects positive for SNP variants. As a finding of this study to test reported SNPs in discrete diabetes population we could find SNPs in new positions in CAPN10 [35], present in some of our study population. The distribution of SNPs among the diabetic population is shown below as in Figure 1 shows the distribution of novel SNP in study population.
We found significant association between CAPN10, ADIPOQ genotypes and metabolic phenotypes. The association of the reported SNPs in Calpain 10 and Adiponectin with type 2 diabetes could be successfully replicated in our discrete study population. A normal individual carrying these gene variants may face greater risk of developing diabetes if having uncontrolled blood sugar levels that exceeds the values of WHO recommendation criterion for diabetes diagnosis.
We conclude that genetic variation in the CAPN10 gene influences blood glucose levels in non-diabetic subjects. This shows the combined effect of more than one factor for outcome of diabetes. The SNP found in new position could be new biomarkers for Type 2 diabetes mellitus. However, further analyses on a larger sample size are required to establish a conclusive association of CAPN10 and ADIPOQ with diabetic population of Andhra Pradesh and Telangana, India.
Download Provisional PDF Here
Article Type: Research Article
Citation: Cheekurhty AJP, Rambabu C, Kumar A (2016) Validation of Reported Single Nucleotide Polymorphisms in the Genes Associated With Type 2 Diabetes Mellitus. J Dia Res Ther 2(1): doi http://dx.doi. org/10.16966/2380-5544.114
Copyright: © 2016 Cheekurhty AJP. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access