Introduction
Subcellular components, directly involved in cancer metabolism, are potentially the most promising biomarkers. Their detection in urine specimens, for example, makes it possible to carry out non-invasive early diagnosis of a variety of urogenital cancers.
Among perspective tumour markers first of all should be mentioned mRNA of the different genes, which are highly expressed during the malignancy processes. For prostate cancer (PCa) the most informative are prostate specific gene PCA3, chimeric gene TMPRSS2-ERG or hTERT gene. Their separate or combined analysis [1-3] yields much more information than the traditional PSA test alone, supplementing at the same time its predictive properties.
The modern approach of gene expression analysis is usually based on the use of quantitative reverse transcription PCR (RT-qPCR) method. Comparing the results with expression data of constitutively expressed reference genes allows monitoring the oscillation of the “specific activity” of the desired gene. However, in routine diagnostics such practices are rare because these methods are first of all very laborious and expensive. In addition, sometimes troubles appear during analysis not only of desired but also of reference genes [4,5].
It would be highly desirable to simplify the methodology of analysis. At least this could be tried to do it during measuring telomerase gene expression [6-8]. Indeed, there was found a strong relationship between hTERT expression in all urogenital cancers or cell lines derived from human PCa and the absence of such relation in healthy tissues [9-11]. This in general allows using a less complex conventional PCR assay for hTERT mRNA analysis, working in a binary “yes/no” classification mode. However, it was recently found, that telomerase activation also takes place in so called “inflammatory” cells. These cells have various natures and accumulate in urine [12]. Consequently, by using telomerase as a cancer marker, it is very desirable to take into account also the source of this enzyme appearance in vivo. Sometimes, this can be done using the capabilities of the real time quantitative PCR method [13].
The aim of this work was to develop a method of measurement of hTERT mRNA levels in urine sediments using another kind of quantitative PCR based on conventional PCR accomplished with ELISA-like detection system (ELOSA - Enzyme-Linked Oligonucleotide Sandwich Assay).
Materials and Methods
Participants
This study (2012-2014) has been approved by the Ethics Committee of Baden-Wurttemberg (Germany). 18 urologists took part in research and provided urine samples of 49 patients with suspected of PCa. Analyses of PSA were also performed in our laboratory. Most of the patients at the time of the survey did not have a definitive diagnosis. In 34 cases the preliminary diagnosis was confirmed with biopsy.
The control group consisted of 32 healthy volunteers not older than 36 years (8 women and 24 men). Some of the young men have turned to the physicians with different problems not associated anyway with prostate disease. Due to their age we have excluded the possibility of PCa existence in this cohort.
All patients gave their written consent to conduct research.
Samples collection and storage
A 50 ml sample of spontaneously voided urine was collected from all probands. The urine samples from men were collected only after a preliminary prostate massage. The delivery of samples to the laboratory was carried out within 24 hours without freezing or any special conditions for transport.
Before starting the collection of the urine sediment, sometimes the samples were placed for several hours at 37°C. In some cases (especially after cooling of the samples) this step was needed to dissolve the sediment in urine.
To obtain cell pellet from urine the so-called fractional centrifugation (3 × 5 min × 400 g) was used, removing the supernatant after each step. After the first centrifugation of original sample (ca. 50 ml) about 35 ml of urine was removed, using a Pasteur pipette connected to a water-jet pump. The remaining urine was stirred vigorously (with Vortex) and transferred to a 14 ml conical tube. After a second centrifugation about 9 ml of urine was removed from the vial in the same manner. The residual urine was vortexed and transferred into 1 ml conical Eppendorf-tube. Only after the last centrifugation, a small “cake” of the cell pellet appeared at the bottom of the test tube. Approximately 900-950 µl of clarified urine was removed, the pellet was re suspended in the remaining volume of urine (about 50- 100 µl) and transferred to 500 µl of Trizol (Ambion). If the volume of sediment suspension exceeded 100 µl we used the more concentrated version of Trizol solution - Trizol LS. The obtained samples were stored before use at -80°C.
Isolation of RNA and cDNA synthesis
Total RNA was isolated from all specimens using the Qiagen RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
Reverse transcription of RNA was done with random primers in a final volume of 20 µl. The reaction mix contained 10 µl purified total RNA, 4 µl 5 × RT buffer solution (Promega), 0,8 µl of 2 mM solution of each dNTPs, 2,2 µl of HOH, 1,0 µl of random primers solution pd(N) 6 (Amersham, 0,26 µg/µl), 1 µl RNase Inhibitor (Promega, 40 U/µl) and 1 µl of M-MLV Reverse Transcriptase (Promega, 200 U/µl). The samples were incubated at 25°C for 10 min, 70°C for 7,5 min, 42°C for 60 min; reverse transcriptase was inactivated by heating at 97°C for 5 min. cDNA was stored at -20°C until being assayed.
PCR Amplification and Detection
We have chosen the beta-2 microglobulin (B2M) gene as housekeeping gene. The levels of B2M mRNA were measured in all cDNA samples with the PCR-method according to [14]. This gene was used as a control for cDNA synthesis indicating that all samples contained cells of different origin. Samples lacking B2M expression were excluded from analysis.
For hTERT mRNA detection we have chosen a method which was used for quantitative analysis of hTERT in snap-frozen tissue or pancreatic juice samples with some modifications [15]. A two-round nested PCR protocol was developed. The assay design involved 15 PCR cycles in the first run (PCR A), predilution of the products (from PCR A) and a second run of 35 cycles (PCR B). The primers and the capture probe are depicted in Table 1. To avoid amplification of contaminating genomic DNA all F- and all R-primers were placed in different exons (GenBank AF015950.1). The PCR with external (F11big & R12big) and internal (F11small & R12small) primers yields a 145-bp and a 88-bp DNA fragment, respectively. Each sample was analyzed using four different cDNA concentrations simultaneously. This was achieved by mixing different amounts of 1/10 diluted cDNA solution (9, 4, 2 and 1 µl), water and master mix, maintaining the constancy of a total volume of the reaction medium (20 µl).
| |
hTERT GenBank AF015950.1) |
Location |
exon |
Product |
|
PCRA |
F11big : 5`- gcg gaa gac agt ggt gaa ct-3`
R12big : 5`- agc tgg agt agt cgc tct gc- 3` |
2758→2777
2903→2884 |
11
12 |
145 |
|
PCRB |
F11small : 5`- tgt aga aga cga ggc cct gg-3`
R12small : 5`-biotin-gta tcc agc agc agg ccg ca-3` |
2782→2801
2870→2849 |
11
12 |
88 |
| Capture Probe |
5`-C12-Aminolink-ctt ttg ttc aga tgc cgg ccc a-3` |
2811→2832 |
11 |
|
Table 1: Oligonucleotide primer and capture probe sequences
In the first PCR run (PCR A) the final reagent concentrations in the reaction mixture were: 1 × PCR-buffer, 0,1% Tween-20, Magnesium chloride - 3,75 mM, 0,2 mM of each dNTPs; enzyme activity, [F11big] & [R12big]-0,4 Units, 375 and 125 nM, respectively. The reaction conditions were: 15 cycles of denaturation at 96°C for 20s, annealing and extension at 70°C for 120s.
The total volume of the second PCR reaction was also 20 µl and involved 18 µl of Master Mix and 2 µl of 1/2000 prediluted product from PCR A. In the PCR B run the final reagent concentrations in the reaction mixture were: 1 × PCR-buffer, 0,1% Tween-20, Magnesium chloride - 2,5mM, 0,2mM of each dNTPs; enzyme activity, [F11small] & [R12small] - 0,1 Units, 50 and 150 nM, respectively. PCR cycling parameters were 10 cycles of 15s at 96°C and 150s at 70°C+10 cycles of 15s at 96°C and 120s at 70°C+15 cycles of 15s at 96°C and 90s at 70°C
PCR was completed with ELOSA detection system. For this purpose a standard DNA binding 96-well microplate (Costar) with immobilized capture probe (10 pmol/100 µl/well) was used. After one hour hybridization at 65°C (5 µl amplicon/100 µl hybridization solution) the plate was treated for 15 minutes at room temperature with a solution of Alkaline Phosphatase conjugated with Streptavidin (Novagen). The progress of the indicator signal was observed by recording the increase of the fluorescence of 4-methylumbelliferyl phosphate after one hour (or more) of reaction time at room temperature.
Results
Purpose and experimental design
We developed a method for the determination of malignant cells in urine sediments. The method is universal for many types of urogenital cancers and is based on the measuring of the hTERT mRNA levels. The main goal of the test was the detection of PCa cells.
Urine sediment collection
In the majority of publications devoted to the use of a variety of urine biomarkers, the main attention usually was focused on a detailed description of the analytical method. The specification of the method for obtaining the urine cell sediment was described as a rule only in common terms, although there is no generally accepted and standardized procedure. However, regarding the heterogeneity of urine consistency, this problem is far from trivial.
Urine, for example, may contain the suspension of insoluble smallest particles. Such particles are able to clog the pores of micron filters suggested to use to receive the cell precipitate. Moreover, sometimes even after a short storage of urine at 4-8°C precipitates are formed. In any case, the method for obtaining the urine sediment is critical for the analysis at a whole and deserves, in our view, a more careful consideration.
The cell pellets were collected from transparent urine samples. If the urine was turbid the vial with the sample was heated for several hours at 37°C with occasional shaking. Often, the turbidity slowly dissolved due to the light warming. The urine sediment was obtained by fractional centrifugation. In most cases, this allowed at the last step of centrifugation to achieve visualization of the precipitate even by the use of completely transparent urine
hTERT mRNA analysis
All urine samples were positive for B2M (data not shown).
For the recognition of cells, expressing telomerase gene we have used the data of four parallel measurements at different initial cDNA concentrations. The results obtained in the study of urine samples of patients with suspicion or confirmed cancer diagnoses are presented in Table 2.
| Diagnosis |
Case/
Internal № |
1 µl cDNA in PCRA |
2 µl cDNA in PCRA |
4 µl cDNA in PCRA |
9 µl cDNA in PCRA |
r |
R2 |
| PCa |
1/65 |
531 |
1611 |
2429 |
2623 |
0,82 |
0,67 |
| PCa |
2/66 |
653 |
510 |
1221 |
1779 |
0,96 |
0,91 |
| PCa |
3/71 |
1181 |
1392 |
1786 |
3125 |
1,00 |
1,00 |
| PCa |
4/72 |
708 |
808 |
1398 |
2585 |
1,00 |
1,00 |
| sus |
5/73 |
468 |
748 |
855 |
1160 |
nd |
nd |
| PCa |
6/74 |
1465 |
1184 |
3174 |
3720 |
0,89 |
0,79 |
| PCa |
7/79 |
449 |
357 |
470 |
803 |
nd |
nd |
| PCa |
8/84 |
1932 |
2338 |
3412 |
4190 |
0,96 |
0,91 |
| PCa |
9/90 |
461 |
629 |
793 |
1218 |
1,0 |
1,0 |
| sus |
10/91 |
696 |
629 |
1953 |
2567 |
nd |
nd |
| PCa |
11/93 |
360 |
391 |
415 |
693 |
nd |
nd |
| PCa |
12/94 |
397 |
369 |
391 |
504 |
nd |
nd |
| PCa |
13/97 |
732 |
1135 |
1022 |
3095 |
0,96 |
0,91 |
| PCa |
14/101 |
1346 |
1428 |
1810 |
2722 |
1,0 |
1,0 |
| sus |
15/102 |
1578 |
1398 |
3479 |
5185 |
nd |
nd |
| sus |
16/103 |
1306 |
1407 |
nd |
3067 |
nd |
nd |
| PCa |
17/104 |
7416 |
8134 |
10542 |
10081 |
0,74 |
0,55 |
| PCa |
18/105 |
2036 |
2402 |
6898 |
6870 |
0,81 |
0,66 |
| PCa |
19/106 |
9327 |
9950 |
10668 |
11851 |
0,98 |
0,97 |
| PCa |
20/107 |
8869 |
9705 |
9647 |
10078 |
0,81 |
0,65 |
| sus |
21/108 |
3409 |
2109 |
5500 |
6711 |
nd |
nd |
| PCa |
22/161 |
665 |
1697 |
1395 |
2634 |
0,89 |
0,80 |
| PCa |
23/164 |
397 |
742 |
1233 |
2191 |
1,00 |
1,00 |
| |
Cut-Off1/2 = 1000/1500 |
|
|
Table 2: Dependence of fluorescence signal on the amount of cDNA - Analysis data of urine samples of patients with suspected and confirmed PCa diagnosis*
*The duration of indicator reaction was 60 min. Abbreviations: nd - not determined, sus - suspicion; r , R2 - the correlation and determination coefficients, respectively
During studying samples from PCa patients, the fluorescence signal, as a rule, was dependent on the initial concentration of cDNA (e.g. Table 2, cases 1/65; 2/66; 3/71; 4/72 etc) and its value was directly proportional to the amount of the cDNA used.
For example, Figure 1 represents the concentration-response curves, received by the examining the samples from several patients with PCa (Table 2, cases 1-4).
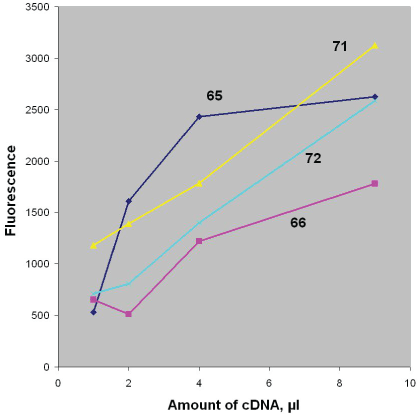
Figure 1: The dependence of fluorescence signal on the amount of cDNA received after proceedings the urine samples from some PCa patients. The internal Nr`s of the patients examined are shown in the figure. (Original data are represented in Table 2, cases 1-4, 60 min duration of indicator reaction)
The results of hTERT mRNA detection obtained during the study of urine samples received from control group probands are presented in Table 3.
| Case/Internal |
1 µl cDNA in PCRA |
2 µl cDNA in PCRA |
4 µl cDNA in PCRA |
9 µl cDNA in PCRA |
r |
R2 |
| 1/92 |
458 |
409 |
458 |
381 |
nd |
nd |
| 2/125 |
2817 |
2006 |
3201 |
2155 |
-0,28 |
0,08 |
| 3/126 |
1181 |
1108 |
2057 |
1141 |
-0,01 |
0,00 |
| 4/127 |
1215 |
665 |
1654 |
797 |
-0,21 |
0,04 |
| 5/128 |
1135 |
629 |
1382 |
726 |
-0,27 |
0,08 |
| 6/129 |
1553 |
1279 |
2487 |
1895 |
0,41 |
0,17 |
| 7/130 |
1520 |
1227 |
2197 |
1993 |
0,63 |
0,39 |
| 8/131 |
366 |
443 |
659 |
504 |
nd |
nd |
| 9/132 |
385 |
385 |
375 |
501 |
nd |
nd |
| 10/133 |
342 |
427 |
409 |
1834 |
nd |
nd |
| 11/134 |
513 |
378 |
476 |
409 |
nd |
nd |
| 12/135 |
595 |
391 |
635 |
482 |
nd |
nd |
| 13/136 |
385 |
427 |
436 |
339 |
nd |
nd |
| 14/137 |
339 |
446 |
476 |
369 |
nd |
nd |
| 15/138 |
351 |
385 |
369 |
400 |
nd |
nd |
| 16/139 |
397 |
586 |
507 |
534 |
nd |
nd |
| 17/140 |
476 |
678 |
629 |
1016 |
nd |
nd |
| 18/141 |
354 |
443 |
482 |
903 |
nd |
nd |
| 19/124 |
394 |
369 |
357 |
598 |
nd |
nd |
| 20/162 |
438 |
421 |
348 |
519 |
nd |
nd |
| 21/163 |
702 |
1257 |
1418 |
2213 |
0,97 |
0,94 |
| 22/165 |
412 |
372 |
433 |
461 |
nd |
nd |
| 23/166 |
534 |
461 |
696 |
1471 |
nd |
nd |
| 24/168 |
449 |
391 |
555 |
571 |
nd |
nd |
| 25/F1 |
574 |
449 |
558 |
836 |
nd |
nd |
| 26/F2 |
501 |
473 |
528 |
803 |
nd |
nd |
| 27/F3 |
507 |
485 |
522 |
632 |
nd |
nd |
| 28/F4 |
485 |
449 |
595 |
543 |
nd |
nd |
| 29/F5 |
568 |
381 |
360 |
464 |
nd |
nd |
| 30/F6 |
418 |
368 |
525 |
427 |
nd |
nd |
| 31/F7 |
394 |
388 |
415 |
690 |
nd |
nd |
| 32/F8 |
400 |
403 |
516 |
409 |
nd |
nd |
| |
Cut-Off1/2=1000/1500 |
|
|
Table 3: Dependence of fluorescence signal on the amount of cDNA -Analysis data of urine samples of probands from control group*
*The duration of indicator reaction was 60 min. Abbreviations: nd - not determined; r , R2 - the correlation and determination coefficients, respectively. The fluorescence data >Cut-Off1 are in bold.
Studying samples from urological patients without PCa or from healthy individuals the measured signals were usually below the Cut-Off (e.g. Table 3, cases 25F1 – 32F8). However, the interpretation of these results was not always unambiguous. In some cases, the measured signals clearly exceeded all Cut-Off values, although they were received from definitely non-cancerous patients (e.g. Table 3, cases 2/125 – 7/130 or Figure 2).
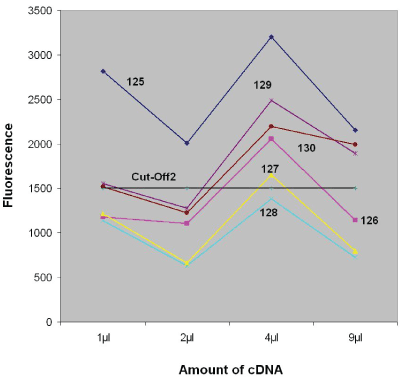
Figure 2: The dependence of false-positive signals on the amount of cDNA received after proceedings the urine samples from some probands of control group without PCa. The internal Nr`s of the patients examined are shown in the figure. (Original data are represented in Table 3, cases 125-130, 60 min duration of indicator reaction)
In addition to the data for the true-positive and false-positive results (Tables 2 and 3) and to the visually depicting of these graphs (Figures 1 and 2) we have also calculated the equations of the trend lines for every case. To estimate how well the regression line represents the experimental data, the corresponding correlation coefficients (r) and coefficients of determinations (R2 ) were calculated (r=√R2 ). Correlations greater than 0.8 are generally described as strong, whereas correlations less than 0.5 are generally described as weak.
By the majority of PCa patients the coefficients of determination observed were equal or greater than 0,8 (strong correlation). The falsepositive results were characterized with a principally another trend: from average (rarely) to very weak correlation (most often). This observation suggests that r or R2 could be used as further parameters for improving the test evaluation. However, the exact determination of a limit values for the coefficients still requires the analysis of a larger number of samples.
Sensitivity and specificity
The preliminary calculation of sensitivity and specificity was made according to standard rules adopted in ELISA and based only on Cut-Off values. The fluorescence magnitudes from blank wells were used for the calculation of Cut-Off1 (2 × Blank) and Cut-Off 2 (3 × Blank). Analytic signal values below Cut-Off 1 were considered as “negative”, above CutOff 2 - as “positive”, and intermediate between Cut-Off 1 and Cut-Off 2 - as borderline. The subsequent changes and additions in the calculation system are discussed in the section “Discussion”.
The data of hTERT mRNA levels in urine samples of 34 patients with a confirmed diagnosis of “Prostate Carcinoma” are represented in the Table 4. According to these results the sensitivity of PCR-ELOSA hTERT mRNA detection method was 71% (borderline results were calculated as negative).
Case/Internal
№ |
Age |
PSA |
Diagnosis, Gleason Score |
hTERT in Urine |
Case/Internal
№ |
Age |
PSA |
Diagnosis, Gleason Score |
hTERT
in Urine |
| 1/65 |
69 |
7,4 |
PCa: 4+4=8 |
pos |
19/110 |
67 |
16,52 |
PCa: 4+5=9 |
pos |
| 2/66 |
71 |
16,89 |
PCa: 3+4=7 |
pos |
19/114 |
62 |
10,38 |
PCa: 4+4=8 |
pos |
| 3/71 |
63 |
13,54 |
PCa: 3+3=6 |
pos |
20/115 |
72 |
5,81 |
PCa: 3+3=6 |
pos |
| 4/72 |
63 |
31,66 |
PCa: 4+5=9 |
pos |
21/116 |
72 |
10,8 |
PCa: 4+4=8 |
pos |
| 5/74 |
72 |
4,6 |
PCa: 4+3=7 |
pos |
21/117 |
62 |
7,73 |
PCa: 3+4=7 |
pos |
| 6/79 |
69 |
13 |
PCa: 3+3=6 |
neg |
23/118 |
66 |
6,52 |
PCa: 3+4=7 |
pos |
| 7/84 |
73 |
13 |
PCa: 4+3=7 |
pos |
24/119 |
70 |
33,1 |
PCa: 4+4=8 |
bd |
| 8/90 |
63 |
4,73 |
PCa: 4+5=9 |
bd |
25/121 |
68 |
8,38 |
PCa: 3+3=6 |
pos |
| 9/93 |
85 |
17 |
PCa: 3+4=7 |
neg |
26/143 |
48 |
10,13 |
PCa: 2+2=4 |
neg |
| 10/94 |
88 |
5,6 |
PCa 3+4=7 |
neg |
27/145 |
72 |
8,56 |
PCa: 3+4=7 |
neg |
| 11/97 |
67 |
6,65 |
PCa 4+3=7 |
bd |
28/153 |
58 |
9,05 |
PCa: 3 +3=6 |
pos |
| 12/104 |
72 |
3,64 |
PCa. 3+4=7 |
pos |
29/155 |
76 |
40 |
PCa: 5+4=9 |
neg |
| 13/105 |
56 |
30,2 |
PCa: 3+3=6 |
pos |
30/156 |
74 |
6,43 |
PCa: 4+4=8 |
neg |
| 14/106 |
71 |
6,8 |
PCa: nd |
pos |
31/159 |
74 |
6,14 |
PCa: nd |
pos |
| 15/107 |
74 |
7,3 |
PCa: 3+3=6 |
pos |
32/160 |
63 |
4,86 |
PCa: 3+3=6 |
pos |
| 16/109 |
75 |
11,46 |
PCa: 4+4=8 |
pos |
33/161 |
66 |
218 |
PCa: 4+4=8 |
pos |
| 17/101 |
66 |
5,12 |
PCa: 3+4=7 |
pos |
34/164 |
67 |
21 |
PCa: 5+4=9 |
pos |
Table 4: The results of hTERT mRNA detection in urine samples of patients with confirmed PCa*
*Abbreviations used: pos - positive; neg - negative; bd - borderline.
The calculation of specificity of the method was based on the results obtained by testing of urine samples from probands of control group (Table 3). The presented data from women were obviously below CutOff 1 at all template concentrations in PCR A / B. Examination of urine samples from men control group showed that there were some results with considerably higher fluorescence values than Cut-Off 2 and therefore must be regarded as “false-positive”. Under these reaction conditions the specificity of the method was ca. 78%.
Discussion
We have presented a method for measuring of telomerase gene mRNA levels in urine sediments. This method is based on conventional nested PCR with ELISA-like detection system (ELOSA) and aimed for the early diagnosis of PCa (or some other urogenital cancers). This technique is well accepted and used for example in a commercial kit for determination of telomerase activity in urine with the same purposes [16,17]. PCRELOSA-tandem proceeding is a low cost alternative to real time RTqPCR. One of the advantages is that the created procedures need only standard PCR- and ELISA-equipment, which is usually available in the most laboratories for molecular biology. Furthermore, the conventional PCR is much simpler and more reliable.
The traditional way of result presentation is another and very important benefit of the used detection system. Indeed, in real time RT-qPCR the results are usually expressed in terms of numbers of amplification cycles (as the rule - threshold cycle values, Ct), but not in terms of concentrations [18]. In contrast, in ELOSA the detectable signals (absorbance or fluorescence) are directly proportional to the analyte concentrations. This easily allows operating with the measured rather than with the calculated parameters and directly observes even a small difference in mRNA concentrations between several samples comparing the intensities of detected signals.
Based on the properties above, it was decided instead of the exact and multiple repetitions of the test to conduct parallel measurements of each cDNA sample at its various concentrations. Indeed, according data of the literature, the results of similar experiments are usually rechecked several times (two or three times).In our case, the evaluation of the end result (positive or negative) will not be based on single or middle values of several signals, but considering them only altogether in a certain relation to one another. In other words, each subsequent measurement must confirm or deny the previous result. This kinetic approach together with the use of Cut-Off ’s values can be seen as an additional stepping-stone for measurement results evaluation. It means, for example, that under the presence of cancer cells in the sample the growth of analytical signal must be expected with the increase of initial concentrations of template in PCRA. In other words, according to Michaelis-Menten enzyme kinetics it must be observed the linear relationship between the fluorescence values and the used amounts of cDNA.
Thus, in the Table 2 are represented the original fluorescence values received by testing the samples from 23 patients with suspicion or confirmed PCa diagnosis. The linear relationship (with correlation coefficients >0,8) between signal values and [cDNA] used was most clearly manifested in more than 90% of all PCa cases.
Recently, it has become more obvious that during hTERT mRNA measurements the inflammatory cells are most often responsible for the appearance of false-positive results. This conclusion was confirmed, in particular, using monoclonal antibodies against hTERT by morphological and immunocytochemical studies of urine cells [12]. The inflammation processes developing due to bacterial infection, urinary stone disease and many other causes are responsible for the emergence in urine of high concentrations of inflammatory cells of different nature [12,18-21]. Thus, to avoid receiving false-positive results, some researchers recommended to perform the hTERT mRNA analysis only after preliminary studies of blood for the presence of activated T-lymphocytes: the samples from patients containing for example activated CD25-positive lymphocytes were excluded from the further investigations [6]. Other authors in such cases had proposed hTERT gene expression analysis only after a prior course of antibiotic therapy [19,22].
So, due to the inflammatory process the concentration of inflammatory cells (e.g. lymphocytes) with activated telomerase increases sharply in urine and therefore in the sample analyzed. This circumstance is one of the reasons of the appearance of “positive” signals from non-malignant cells. From the viewpoint of the purpose of analysis these results should be considered as false-positive. For example, by the testing of urine samples from control group probands there were received 9 potentially positive results (Table 3; cases 2/125-7/130, 10/133; 21/163 and 23/166). According to physician anamnesis, in 5 cases (Table 3; cases 2/125, 6/129, 7/130, 21/163 and 23/166) the samples were obtained from probands with urogenital tract inflammation
Of course, the amount of inflammatory cells in urine will depend on the strength of inflammation. However, in those cases the number of corresponding non-malignant cells with active telomerase will be more often significantly exceed the potential number of cancer cells at the initial stage of PCa. Just, due to the excessive amount of hTERT mRNA from inflammatory cells being analyzed, under the indicated reaction conditions the linear relationship between the magnitude of the signal and corresponding [cDNA] most likely is absent. Indeed, as it could be seen from the presented data in most cases (Table 3; cases 2/125 - 7/130) the dependence of “false-positive” signals vs. [cDNA] has a haphazard distribution (Figure 2). This made us to suppose, that the absence of reliable correlation between the initial cDNA concentrations and the values of analytical signals exceeding Cut-Off could be a characteristic feature of false-positive results.
Thus, the analysis of concentration-response curves can provide additional criterions for the evaluating of the nature of the «positive» signal (>Cut-Off). The directly proportional relationship between the initial concentrations of cDNA and signal values should be observed, as a rule, in the presence of cancer cells in the sample. The absence of such dependence or the haphazard analytical signals (>Cut-Off) distribution vs [cDNA] could be indicate the presence in the sample of a large number of inflammatory cells. Of course, this is not always the case (see e.g. Table 3, case 21/163). However, maybe this time the observed linear dependence between signal values and cDNA concentrations (which completely imitates the pattern obtained during PCa samples studies) could simply be explained by the presence of a mild inflammation, and this is the case where the exception confirms the rule.
Moreover, it should be noted that not only the above-mentioned “medical indications” or so-called “urothelial conditions” [3,23], but also some of the imperfections of the test itself or even slight changes in the reaction conditions are responsible for the appearance of falsepositive results. For instance, the reaction mixture for PCRB must not be “overloaded” with DNA fragments after PCRA: 10-fold reduction in preliminary dilution of the intermediate PCR-product (1/200 instead of 1/2000) was resulting in the growth of number of false-positive results by more than two times (an increase of 100%), while the number of true-positive results was changing by no more than about 10% (data not presented). Thus, owing to the different nature of the appearance of false-positive results, the proposed approach of their evaluation cannot be comprehensive. At the same time, this principle allowed us, in particular, to regard the results of 6 out of 9 potentially “positive” measurements (Table 3, cases 2/125-7/130) as false-positive. In this case, the recalculated specificity of the method increases from 78 to 91%.
Conclusion
We describe a method for determining the levels of hTERT mRNA. The method is designated for the detection of cancer cells in urine sediments and verified by using the samples from patients with prostate cancer. The quantitative measurement of the PCR products was done with an ELISA-like technique. The main feature of the method is the simultaneous conducting of parallel experiments with several various cDNA concentrations. Respectively, the evaluation of the end result (positive or negative) will be based on considering all measurements altogether with a certain relation to each other. It is supposed to use the revealed type of correlation between initial cDNA concentrations and fluorescence values as a template to recognize the nature of signals which exceed the Cut-Off. This approach together with the use of Cut-Off values can be seen as an additional stepping-stone for evaluation of hTERT mRNA measurements. Based on these principles the sensitivity and specificity of the represented method are 71% & 91%, respectively. The proposed method made it possible to distinguish in many cases the true- and false-positive results without measuring the ratio of hTERT/reference gene mRNA levels. The detection of housekeeping gene is only needed to confirm the successful isolation of urine cells and to prove the quality of extracted genetic material for the amplification.
Acknowledgement
The authors thank Dr. Karl-Ernst Ambs, Dr. Peter Dorner, Dr. Markus Hack, Dr. Dieter Egle, Dr. Samir Salameh, Dr. Christoph Gudemann, Dr. Joachim Felgner, Dr. Roland Britzelmaier, Dr. Gerhard Rimmelspacher and other physicians, who have provided the clinical and supplementary material for this study. The authors thank our colleagues David Seiller, Dr. Klaus Schneider, Dr. Hennrik Schroeter, Dr. Beate Boldrin and Anke Colberg for technical assistance and useful discussion.