Introduction
Johne’s disease, caused by Mycobacterium avium subspecies paratuberculosis (MAP), is being increasingly recognized as a significant herd health problem; thus, there is a need for a reliable diagnostic tool for large-scale use to facilitate control and prevention programs and eventually eradicate the disease. MAP is an extremely slow-growing, acidfast and mycobactin-dependent pathogen to grow. Infection with this bacterium leads to a chronic granulomatous enteritis in a broad range of hosts mainly in cattle and other wild and domestic ruminants [1].
Detection of MAP-infected animals poses great difficulties, thus rapid and reliable procedures for the direct detection of MAP in the herd and food samples are still challenging. Efforts have been made in the last few decades to develop protocols for the detection of MAP in feces, milk, tissue, food, and environmental samples using various methods. Fecal culture and serology are the most commonly used tests in the clinical laboratories [1]. The gold standard for Johne’s disease is identification of the etiologic agent, MAP, in tissues that show characteristic histopathologic lesions [2]. Culture will require 8 to 12 weeks and has well known limitations in sensitivity, especially in sub clinically infected cattle, and turnaround time; nonetheless, culture or some other method to detect MAP directly still has a place at certain stages of a Johne’s disease control program [3].
With the development of new molecular techniques, more powerful protocols have become available to improve the detection of MAP. PCR is a powerful method for specific detection of DNA sequences, for which samples can be taken from colostrum, milk, feces, and tissues [4,5]. For molecular detection of MAP, PCR assays targeting various regions specific for MAP were reported from different laboratories. Some of the PCR target genes include MAP-specific genomic sequences such as the insertion sequence IS900, F57 element, IS1311 insertion sequence and hsp X gene [5-7]. Of these, multi-copy IS900 is a target of choice and is the most widely used in the characterization of MAP. The characterization of IS900 insertion sequence, which has 1,451 base pairs and is present as 15 to 20 copies in the MAP genome, has enabled the accurate identification of minimum amounts of bacterial DNA by the polymerase chain reaction (PCR) assay [8]. Furthermore, by suppression subtractive hybridization-based method, nine novel MAP fragments of between 318 and 596 bp have been identified and characterized. A database search revealed little or no similarity with other Mycobacterium species [9].
It is essential to authenticate the existing protocol and targets to have an efficient method for the detection of M. avium subsp. paratuberculosis. The objective of this study was to compare and validate the real-time PCR results of IS900-insertion sequence and nine putative sequences as diagnostic tools to detect MAP.
Materials and Methods
Bacterial strains and growth
Three M. avium subsp. paratuberculosis strains, (baa-968, 19851 and 43015), six non-MAP species (M. fortuitum subsp. fortuitum, M. intracellulare, M. kansasii, M. marinum, M. phlei, and M. scrofulaceum) and 13 MAP clinical isolates were included in this study. The MAP strains and the non-MAP species were purchased from American Type Culture Collection (ATCC, Manassas, VA). The clinical MAP strains, isolated from cattle feces with naturally acquired Johne’s disease, were obtained from Thompson-Bishop-Sparks State Diagnostic Laboratory, Auburn, Alabama. The MAP was grown in Lowenstein-Jensen media (Becton Dickinson and Company, Sparks, MD) and 7H9 broth supplemented with 2 mg of Mycobactin (Difco Laboratories, Detroit, MI).
DNA Extraction
PrepMan kit (Applied Biosystems, Carlsbad, CA) was used to extract DNA from the culture. Briefly, the samples were centrifuged at a medium speed (3 min at 10,000 × g) on a tabletop centrifuge to pellet the cells. After the supernatant has been discarded, the pellet was washed and resuspended in 100 µl of the PrepMan reagent. After brief vortexing, the mixture was boiled at 100°C for 10 min. The preparation was centrifuged, and the supernatant was used as the template for PCR and stored at -20°C for future use. Before PCR assay, the extracted DNA was quantified by ND-1000 spectrophotometer (NanoDrop, Wilmington, DE).
Real-time Polymerase Chain Reaction
The sample preparation, DNA extraction, PCR mixture assembly, and post-PCR analysis were performed in separate laboratory rooms. DNA extracted from the culture was quantified and subjected to real-time PCR. The real-time PCR assay was performed using Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technologies, Santa Clara, CA). The PCR targets were IS900 insertion sequence and other nine putative sequences [AF503872 (Mptb52.1), AF503873 (Mptb52.16), AF503874 (Mptb52.6), AF503875 (Mptb54.16), AF503876 (Mptb54.24), AF503877 (Mptb54.33), AF503878 (Mptb55.15), AF503879 (Mptb61.32), AF503880 (Mptb62.12)] described and deposited in GenBank [9,10].
Primers specific for the targeted genes (Table 1) were selected from previously published literature and optimized. Controls consisted of reaction mixture alone (negative control) and a positive control containing one µl of genomic DNA from reference M. avium subsp. paratuberculosis (ATCC baa 968, 19851 and 43015). Melting curve profile and conventional gel analysis were performed to confirm the positive PCR results. All realtime PCR assays were performed at least in duplicate using Stratagene Mx3000P® real-time PCR instrument (Stratagene, La Jolla, CA).
| Accession (definition) |
Primer sequence (5’-3’) |
Amplicon size (bp) |
Start -End position: |
References |
| AF503872 (Mptb52.1) |
F- ATCCGGTGGATCGTCTCGC
R- CATCGATTGACGTCCTCACC |
371 |
3332937-3333307 |
[9] |
| AF503873 (Mptb52.16) |
F- ACGACAACGACCACACTCC
R- CACAGGACACTCCATCAGG |
396 |
3907417-3907812 |
[9] |
| AF503874 (Mptb52.6 ) |
F- ATCAACAAGACGTGCGCG
R- CAGTGGTGCGCATGAAGTC |
266 |
3327332-3327597 |
[9] |
| AF503875 (Mptb54.16) |
F- CTTCAACTCGACAAGAGTCGT
R- TCAGAAGCGCGACTGGACTT |
403 |
3324756-3325158 |
[9] |
| AF503876 (Mptb54.24) |
F- GCATGAACAGCAACAGGACC
R- TGCGGTGGTAGCAACTAGCC |
410 |
1823028-1823437 |
[9] |
| AF503877 (Mptb54.33) |
F- GATCTCTACGTGATCAAGGAG
R- CGTCGGACAGGCGTATGTAA |
415 |
1823550-1823964 |
[9] |
| AF503878 (Mptb55.15) |
F- TTGGACGAACTTGGCGATGA
R- GGTAATCGGAACCCTTAGCG |
546 |
1825825-1826370 |
[9] |
| AF503879 (Mptb61.32) |
F- ACGACGTCGTCAGACACCTA
R- CGTAGGGTATATCGGGTCAAG |
523 |
29424-29946 |
[9] |
| AF503880 (Mptb62.12) |
F- CTTGATCCTCGGGAGCGTG
R- CTCCGACTGAATGAGCTGTG |
474 |
4262495-4262968 |
[9] |
| mHM172613 (IS900) |
F- CCGCTAATTGAGAGATGCGATTGG
R-AATCAACTCCAGCAGCGCGGCCTCG |
229 |
4747310-4747538 |
[10] |
| F- forward primer and R- reverse primer. |
Table 1: Specific primers used in the real-time PCR to amplify IS900 and the putative sequences with the corresponding amplicon size.
Gel electrophoresis
After PCR was completed, 10 µl of the real-time PCR amplification reaction was mixed with GelRed (Biotium, Hayward, CA) mixture and electrophoresed in Tris- acetate-EDTA buffer through a 2% agarose gel. Bands of the appropriate size were identified by comparison with a DNA ladder (Roche Molecular Biochemicals, Indianapolis, IN).
In silico PCR
In silico PCR (virtual PCR) was performed from a sequence database with selected pairs of PCR primers, using an indexing strategy for fast performance. The virtual PCR is considered to have high stringency [11].
The specificity of the primers was confirmed by in silico PCR Amplification against 65 Mycobacterium species. In silico PCR searches a sequence database with a pair of PCR primers, using an indexing strategy for fast performance. This PCR simulation was performed against the most current sequenced Mycobacterium genomes which allow a maximum of 2 mismatches between primers and templates, so the stringency of in silico PCR must be considered high. All the currently available Mycobacterium species sequence data (Table 2) were checked against the selected primers.
| S.No |
Mycobacterium Species |
Strains |
| 1. |
Mycobacterium abscessus |
ATCC 19977 |
| 2. |
Mycobacterium abscessus subsp. bolletii |
50594 |
| 3. |
Mycobacterium africanum |
GM041182 |
| 4. |
Mycobacterium avium |
104 |
| 5. |
Mycobacteriumaviumsubsp.paratuberculosis |
MAP4 |
| 6. |
Mycobacterium avium subsp. paratuberculosis str. |
k10 |
| 7. |
Mycobacterium bovis BCG str. |
Korea 1168P |
| 8. |
Mycobacterium bovis BCG str. |
Mexico |
| 9. |
Mycobacterium bovis BCG str. |
Pasteur 1173P2 |
| 10. |
Mycobacterium bovis BCG str. |
Tokyo 172 |
| 11. |
Mycobacterium bovis subsp. bovis |
AF2122/97 |
| 12. |
Mycobacterium canettii |
CIPT 140010059 |
| 13. |
Mycobacterium canettii |
CIPT 140060008 |
| 14. |
Mycobacterium canettii |
CIPT 140070008 |
| 15. |
Mycobacterium canettii |
CIPT 140070010 |
| 16. |
Mycobacterium canettii |
CIPT 140070017 |
| 17. |
Mycobacterium chubuense |
NBB4 |
| 18. |
Mycobacterium gilvum |
PYR-GCK |
| 19. |
Mycobacterium indicus pranii |
MTCC 9506 |
| 20. |
Mycobacterium intracellulare |
ATCC 13950 |
| 21. |
Mycobacterium intracellulare |
MOTT-02 |
| 22. |
Mycobacterium intracellulare |
MOTT-64 |
| 23. |
Mycobacterium kansasii |
ATCC 12478 |
| 24. |
Mycobacterium leprae |
Br4923 |
| 25. |
Mycobacterium leprae strain |
TN |
| 26. |
Mycobacterium liflandii |
128FXT |
| 27. |
Mycobacterium marinum |
M |
| 28. |
Mycobacterium massiliense str. |
GO 06 |
| 29. |
Mycobacterium neoaurum |
VKM Ac-1815D |
| 30. |
Mycobacterium rhodesiae |
NBB3 |
| 31. |
Mycobacterium smegmatis |
JS623 |
| 32. |
Mycobacterium smegmatis str. |
MC2 155 |
| 33. |
Mycobacterium smegmatis str. |
MC2 155 |
| 34. |
Mycobacterium sp. |
JDM601 |
| 35. |
Mycobacterium sp. |
JLS |
| 36. |
Mycobacterium sp. |
KMS |
| 37. |
Mycobacterium sp. |
MCS |
| 38. |
Mycobacterium sp. |
MOTT36Y |
| 39. |
Mycobacterium sp. |
Spyr1 |
| 40. |
Mycobacterium tuberculosis |
7199-99 |
| 41. |
Mycobacterium tuberculosis |
CAS NITR204 |
| 42. |
Mycobacterium tuberculosis |
CCDC5079 |
| 43. |
Mycobacterium tuberculosis |
CCDC5079 |
| 44. |
Mycobacterium tuberculosis |
CCDC5180 |
| 45. |
Mycobacterium tuberculosis |
CDC1551 |
| 46. |
Mycobacterium tuberculosis |
CTRI-2 |
| 47. |
Mycobacterium tuberculosis |
EAI5 |
| 48. |
Mycobacterium tuberculosis |
EAI5 NITR206 |
| 49. |
Mycobacterium tuberculosis |
F11 |
| 50. |
Mycobacterium tuberculosis |
H37Ra |
| 51. |
Mycobacterium tuberculosis |
H37Rv |
| 52. |
Mycobacterium tuberculosis |
H37Rv |
| 53. |
Mycobacterium tuberculosis |
KZN 1435 |
| 54. |
Mycobacterium tuberculosis |
KZN 4207 |
| 55. |
Mycobacterium tuberculosis |
KZN 605 |
| 56. |
Mycobacterium tuberculosis |
RGTB327 |
| 57. |
Mycobacterium tuberculosis |
RGTB423 |
| 58. |
Mycobacterium tuberculosis |
UT205 |
| 59. |
Mycobacterium tuberculosis str. Beijing |
NITR203 |
| 60. |
Mycobacterium tuberculosis str. Erdman |
= ATCC 35801 DNA |
| 61. |
Mycobacterium tuberculosis str. Haarlem |
|
| 62. |
Mycobacterium tuberculosis str. Haarlem |
NITR202 |
| 63. |
Mycobacterium ulcerans |
Agy99 |
| 64. |
Mycobacterium vanbaalenii |
PYR-1 |
| 65. |
Mycobacterium yongonense |
05-1390 |
Table 2: Mycobacterium strains validated using virtual in silico PCR amplification. The corresponding primers are listed in Table 1.
Results and Discussion
Availability of the completed sequences of MAP genome supported the identification of more specific diagnostic sequences [12,13]. The availability of the sequence database brings clarity to unresolved questions surrounding the pathogenesis of MAP in animals and humans and also facilitates the development of detection kits. The introduction of PCR targeting repetitive sequences like IS900 insertion element, which is present in multiple copies in MAP genome, has been reported to reduce the time and labor required for the MAP detection [10]. The primary focus of this study was to evaluate and optimize the specificity of PCR-based molecular detection for the MAP targeting insertion sequence IS900 and other repetitive sequences as uniplex and/or multiplex PCR to detect MAP. To validate the IS900 insertion sequence and the repetitive/putative sequences, appropriate primers were selected/adopted and validated by both real-time PCR and in silico PCR.
Real-time PCR
In this study, the specificity of the real-time PCR assays was optimized by the analysis of genomic DNA from a panel of different Mycobacterium species which includes three MAP reference strains, 13 MAP clinical isolates from cattle and six non-MAP isolates. The genomic DNA samples were subjected to PCR assays targeting IS900 insertion sequence and additional nine putative sequences against MAP and non-MAP cultures as uniplex and also multiplex platforms. The results showed that IS900 and the putative sequence-specific primer pairs were amplified only from genomic DNA extract of the MAP reference strains and clinical MAP isolates. As shown in Figure 1, there is no amplification from the nonMAP strains (M. fortuitum subsp. fortuitum, M. intracellulare, M. kansasii, M. marinum, M. phlei, and M. scrofulaceum). As demonstrated on the gel, the amplicons from MAP were at the expected molecular weight and no signals were detected for the non-MAP species (Figure 2). Uniplex and/ or multiplex-PCR of IS900 insertion sequence and the putative sequences showed the presence of single or two distinct amplicons, respectively (Figure 2). The lower band represented the IS900 sequence amplicon (229 bp), and the top band represents amplicon from Mptb52.1 (371 bp).
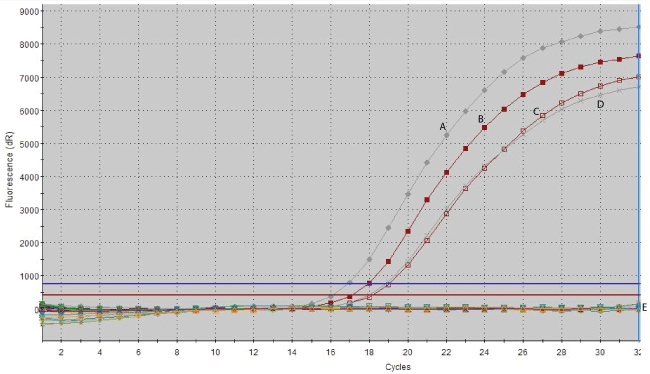
Figure 1: Real time PCR of a unique IS900 target sequence amplified from two M. avium subsp. paratuberculosis reference strains (A: ATCC baa-968; B : 43015 B), clinical MAP isolates (C and D) and E: non-MAP species (M. fortuitum subsp. fortuitum, M.intracellulare, M. kansasii, M. marinum, M. phlei, and M. scrofulaceum).
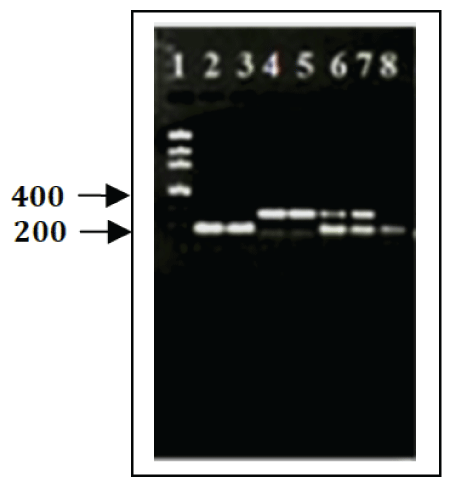
Figure 2: PCR amplification of IS900 and Mptb52.6, putative sequence, from M. avium subsp. paratuberculosis.. PCR products were subjected to 1.5% agarose gel electrophoresis and amplification products were verified by gel electrophoresis. (A) Lane 1, Low DNA Mass Ladder; lanes 2, 3 and 8 PCR product of IS900 Insertion sequence; lanes 4 & 5, PCR product of Mptb52.6; lanes 6 & 7, amplifications of double-PCR product of targeting IS900 & Mptb52.16 sequences.
In silico PCR analysis
This virtual PCR was simulated against up-to-date sequences of Mycobacterium genomes in the database. A total of 23 Mycobacterium spp and 65 strains were verified against IS900 insertion sequence and the other nine repetitive sequences (Table 2). The virtual PCR results showed that IS900 and the putative sequence–specific primer pairs amplified specific targets only from MAP. Furthermore, the in silico simulation results showed that all the primers selected for each of IS900 segment and the putative sequences were amplified only from MAP samples and not from any of the non-MAP species. The amplicon sizes were at the expected range as in published literature and no secondary bands were detected (Figure 3).

Figure 3: In silico PCR amplification results (10 target primers against a total of 65 MAP and non-MAP mycobacterial strains. Mycobacterium strains listed as number 5 (Mycobacterium avium subsp. paratuberculosis MAP4) and number 6 (Mycobacterium avium subsp. paratuberculosis str. k10) were amplified (Refer Table 2 for the Mycobacterium spp. list).
In general Several PCR assays have been developed for the detection of M. avium subsp. paratuberculosis [14,15]. The most prominent target used in several studies to detect DNA of M. avium subsp. paratuberculosis by PCR is the insertion element IS900 [15-17]. The multicopy (17 copies) nature of the IS900 sequence on the M. avium subsp. paratuberculosis chromosome makes it ideal as a target sequence for the detection of MAP, since it exhibits a higher level of sensitivity compared to the use of singlecopy genes as targets [18,19].
The specificity of IS900 diagnostic PCR, however, has been also questioned and some reports indicated the presence of IS900-like sequences in Mycobacterium species other than MAP which may increase the risk of false MAP positive results [20,21]. Rajeev et al. (2005) reported the isolation of one non-MAP Mycobacterium strain positive for IS900 PCR, which has not yet been typed [22].
Some reports described that the problem of false negative PCR results might arise from difficulties in the disruption of the robust cell wall of MAP and insist that the mechanical disruption step may be needed to ensure the maximal release of DNA from MAP cells present in the tissue sample. The efficient procedure to release the DNA could hence maximize the PCR detection sensitivity [10].
In general, the diagnostic performance of IS900 insertion sequencetargeted detection techniques could be limited or misinterpreted due to various factors. These include isolation of organism-specific DNA; origin of the sample; lack of selection of proper primers; PCR amplification in the presence of inhibitory substances in preferred clinical specimens; and also the presence of concurrent Mycobacterium species [1,15]. Furthermore, the possible explanations for this heterogeneity might be host-pathogen interactions, host preference of MAP strains and pathogen adaptation to the environmental factors [10]. Based on the samples tested and the methodology outlined in our study, IS900 and the additional nine putative sequences are specific to use in PCR as uniplex and/or multiplex applications.
Acknowledgement
This work was supported by U.S. Department of Agriculture Grant (USDA/CRIS, Accession 0203750) and USDA/NIFA (GRANT#2011-51110-31082). We would like to thank Dr. Sircy Moore and the bacteriology staff of the Thompson-Bishop-Sparks State Diagnostic Laboratory, Auburn, AL. We acknowledge Dr. David Nganwa for the editing support and Tuskegee University’s College of Veterinary Medicine (CVM) Center of Excellence (Grant #245 5D34HP00001-20-00) for Tuskegee University Shared Instrumentation Core Facility is supported by NIH/NIMHD grant G12MD007585. The summer research support.
Article Information
Article Type: Research Article
Citation: Yehualaeshet T, Graham M, Samuel T, Morton SER, Rodning S, et al. (2017) Comparison and Validation of IS900-Sequence and Putative Sequences as Specific Target to Detect Mycobacterium avium subsp. paratuberculosis. J Anim Sci Res 1(1): doi http://dx.doi.org/10.16966/2576-6457.101
Copyright: © 2017 Yehualaeshet T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
Received date: 25 May 2017
Accepted date: 17 Jul 2017
Published date: 24 Jul 2017




