Abstract
Methicillin resistant and susceptible Staphylococcus aureus (MRSA and MSSA respectively) remain a public health concern as human
pathogens. Presence of MRSA and MSSA in river water and urban effluents was studied to analyze the S. aureus population and determine the
genetic diversity and predominant genotypes obtained by spa types and MLST on each ecological niche. MRSA proportion in urban effluents
was higher than in river water (P<0.05). According to the Simpson’s Index of Diversity based on spa types, MSSA isolates were more diverse
than MRSA isolates (P<0.05). Predominant spa types and STs detected in MSSA river water isolates were different from those found in urban
effluents. In the MRSA population, ST125-t067 was the predominant genotype detected in both urban effluents (67.6%) and river water (82.4%).
Overall, the MSSA and MRSA lineages most frequently found in river water and urban effluents were human associated clones (ST125-t067,
ST5-t002; ST22-t032, ST30-t012 and ST15-t084). These results show the potential role of water in the S. aureus maintenance and dissemination.
Association of isolates from the river with human ones could be reflecting the effect of anthropogenic activities in the ecosystems, which highlights
the need to evaluate the circulation of pathogens in the environment via water.
Keywords
Staphylococcus aureus; Urban effluents; River water; Spa typing; MLST
Introduction
Methicillin resistant and methicillin susceptible Staphylococcus aureus
(MRSA and MSSA) remains a public health concern as human pathogens
[1]. Different genetic lineages have been described as Hospital AssociatedMRSA
(HA-MRSA), Community-Associated-MRSA (CA-MRSA) and
Livestock Associated-MRSA (LA-MRSA). Infections caused by HAMRSA
isolates are normally related to risk factors such as hospitalization,
surgery or indwelling medical devices [2]. CA-MRSA affects to otherwise
healthy people and infections have been linked to the presence of the
toxin Panton-Valentine leukocidin or PVL [2]. Finally, LA-MRSA has
been considered an occupational risk although its frequency of isolation
is increasing in countries with low prevalence of MRSA [3]. Genetic
differentiation between these groups is getting more complicated due
to the incidence of HA-MRSA in the community and vice versa and
due to the transmission of MRSA between humans and animals [4].
Direct contact was pointed out as the most feasible transmission route
of S. aureus [3]. However, colonized individuals might discharge bacteria
into urban effluents and recreational water [5,6]. Wastewater treatment
plants have been described as reservoirs for MRSA, and hypothetically,
participate in their dissemination through sewage treatment plant
effluents, as part of the S. aureus population might survive the wastewater
treatments [6-9]. Moreover, the presence of MRSA in river water [10]
points out the potential role of water in the dissemination of MRSA, and
in consequence, into associated environments [8, 9, 11]. These facts led
us to study the presence of MSSA and MRSA in urban effluents and river
water to assess the genetic diversity and predominant genotypes within
each ecological niche.
Experimental Section
Samples origin
One sample of urban effluents was taken in July 2011 in a sewage
plant that gathers wastewater from several urban collectors (untreated
wastewater) in an urban nucleus with 3.2 million people (http://www.ine.
es/SID/Informe.do). One river water sample was taken in September 2012
in the countryside, downstream the municipal term of a city with 8,392
people (http://www.ine.es/SID/Informe.do).
Isolation and characterization
Both samples were divided into sub-samples and processed separately
(n=100 subsamples per sample). Each sub-sample (1 mL) was cultured
on 9 mL of Muller-Hinton broth (6.5% NaCl, Oxoid) and incubated at
37°C for 16-20 h. One mL was then transferred to 9 mL tryptone soy
broth (Oxoid) with cefoxitin (3.5 mg/L, Sigma–Aldrich) and aztreonam
(75 mg/L, Sigma–Aldrich) and incubated at 37°C for 16–20 h. Finally,
25 µL were streaked onto Brilliance MRSA plates (Oxoid) and incubated
for 24–48 h at 37°C [10]. Denim blue colonies (one per subsample) were
confirmed as MRSA (mecA or mecC positive) by PCR [12]. In parallel,
100 µL of incubated Muller-Hinton broth (6.5% NaCl) were cultured
onto Baird Parker (BP) agar with Rabbit Plasma Fibrinogen (bioMerieux)
and incubated at 37°C during 24-48 h. Black colonies coagulase-positive
(one per subsample) were selected as potential S. aureus and confirmed
as MRSA (mecA or mecC positive) or MSSA (mecA and mecC negative)
as described above. Confirmed S. aureus were characterized by spa typing
sequencing the variable fragment of protein A [12], and spa types were
analysed by the minimal Spanning tree algorithm (Bionumerics 6.0).
Simpson’s Index of Diversity (SID) and Jackknife pseudo-values (CI: 95%)
were used to estimate the genetic diversity of S. aureus isolates based on
spa types (Figure 2; http://darwin.phyloviz.net/ComparingPartitions/
index.php?link=Tool). Multilocus Sequence Typing (MLST) was
performed to at least one isolate per spa type and isolation route
(n=103) to obtain the sequence types (STs) according to the protocol
described before [13]. Detection of Panton–Valentine leukocidin (PVL)
was also carried out [12].
Statistical analysis
Fisher’s exact test (SPSS 20) was calculated to analyze the relationship
between the type of sample (urban effluents or river water) and the
presence of MRSA and between the type of sample and the most frequent
spa types and STs in the collection (n>5 isolates).
Results and Discussion
MRSA protocol detected 96 MRSA isolates out of 100 subsamples in
urban effluents, meanwhile only 33/100 MRSA in river water (Table 1). All
isolates obtained by this protocol were mecA-MRSA. On the Baird Parker
protocol, most of S. aureus isolates were MSSA (Table 1), but some MRSA
were also detected (5 isolates mecA-MRSA and 1 isolate mecC-MRSA in
urban effluents and 1 mecA-MRSA in river water). The low detection rates
of mecC-MRSA compared with mecA-MRSA is in agreement with other
studies [13-16]. However, the detection of mecC-MRSA in water effluents
is of interest considering the potential for zoonotic transmission [17] and
wildlife-environmental interactions of mecC-positive MRSA [10].
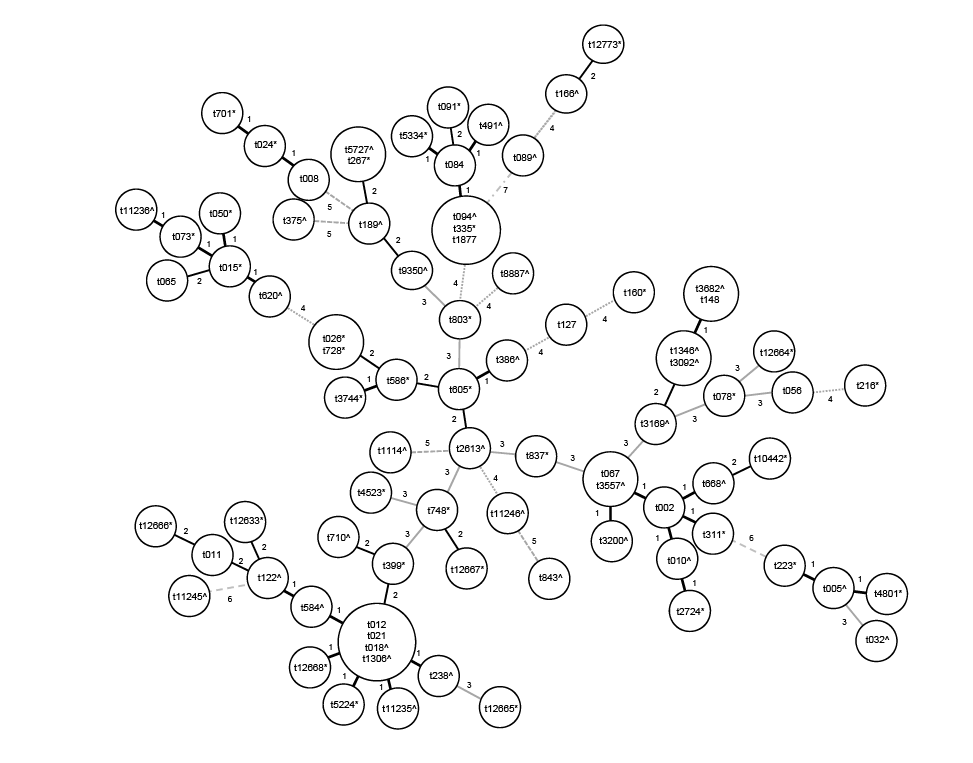
Figure 1: Clustering of spa types by minimal Spanning tree algorithm. Lines/numbers between circles represent the genetic distance between
different spa-types. (*): spa types detected in river water; (^): spa types detected in urban effluents and ( ): shared spa types
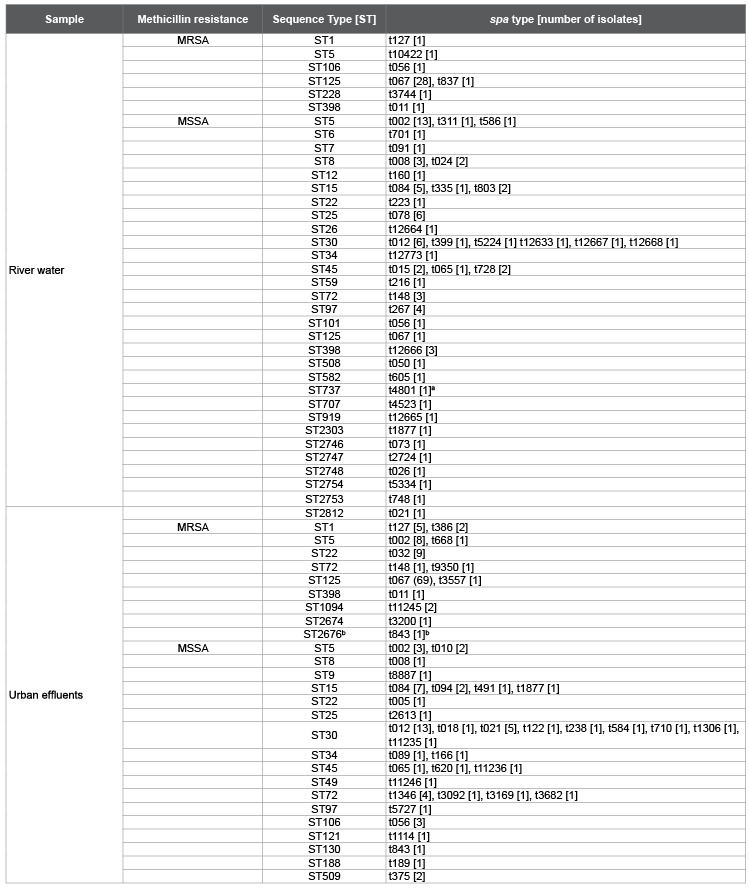
Table 1: Spa types and sequence types [STs] of S. aureus found in river water and urban effluents.
MRSA: methicillin resistant S. aureus; MSSA: methicillin susceptible S. aureus; a
PVL positive isolate; b
mecC isolate [13]; bold letter: spa types and STs
described in this study
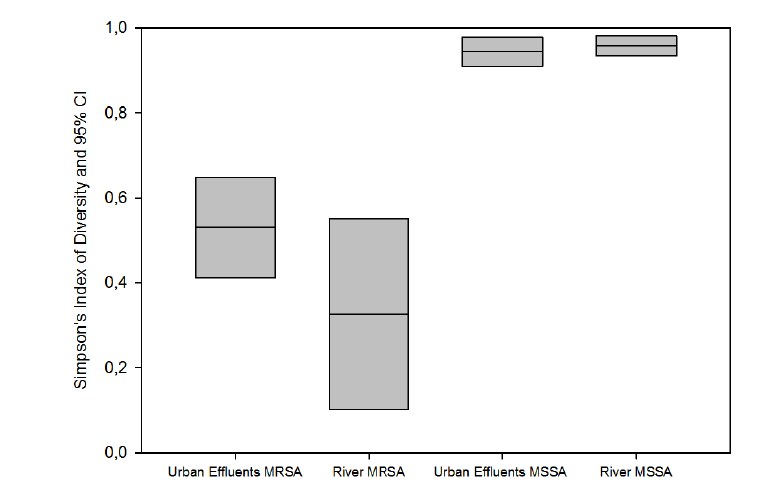
Figure 2: Genetic diversity of S. aureus isolates based on spa types: Simpson’s Index of Diversity (Simpson’s ID) and Jackknife pseudo-values
confidence intervals (CI) at 95%. MRSA: methicillin resistant S. aureus; MSSA: methicillin susceptible S. aureus
Only one isolate MSSA from river water was positive to PVL (ST737-
spa type t4801). Some studies described that PVL is increasing in the south
of Europe and in some areas in Spain, but those are referred mainly to ST8
and ST80 [18,19], STs whose isolation frequency in our study (Table 1)
was low (ST8) or undetected (ST80).
A higher proportion of MRSA isolates was detected in urban effluents
(102/169; 60.4%) than in river water (34/115; 29.6%), differences
statistically significant (P<0.05). This higher frequency of isolation of
MRSA in urban effluents compared with the river water might be related to
the higher concentration of antimicrobial resistant bacteria in wastewater
and the population density in the area of sampling [20,21].
Isolates were grouped in 81 different spa types (Figure 1) and 42 STs,
with 12 spa types and 14 STs being common to both environments (Table
1). Ten new spa types and 7 new STs were firstly described in this study
(Table 1). Forty-two different spa types were detected in MSSA isolates
from river water and 35 in urban effluents. On MRSA isolates, the number
of different spa types detected from river water and urban effluent were
7 and 13, respectively (Table 1). This genetic diversity observed in the
bacterial population of MSSA and MRSA isolates would reflect the S.
aureus population in both water samples.
MSSA were genetically more diverse than MRSA isolates (Figure 2;
P<0.05). Simpson’s Index of Diversity (SID) based on spa types was 0.958
(95% CI: 0.934-0.981) for MSSA isolates from river water and 0.944
(95% CI: 0.910-0.978) for MSSA isolates from urban effluents (Figure 2;
P>0.05). This genetic diversity observed in MSSA isolates is similar to that
observed in previous studies [1]. Despite this high genetic diversity, some
MSSA genotypes were more frequently isolated. Thus, the most frequent
MSSA genotypes detected in river water were ST5/spa type t002 (13/81;
16.0%), ST30/spa type t012 (6/81; 7.4%), ST25/spa type t078 (6/81; 7.4%)
and ST15/spa type t084 (5/81; 6.2%), while ST30/spa type t012 (13/67;
19.4%), ST15/spa type t084 (7/67; 10.4%) and ST30/spa type t021 (5/67;
7.5%) were the most frequent MSSA genotypes in urban effluents. Some
of these frequent MSSA genotypes have been previously identified in
human healthy carriers and patients [1,22,23].
Regarding MRSA isolates from river water and urban effluents, SID
values were 0.326 (95% CI: 0.102-0.550)) and 0.530 (95% CI: 0.412-
0.648) respectively (P>0.05).This low genetic diversity is due to the
existence of predominant genotypes that included most of the MRSA
isolates. In particular, the genotype ST125/spa type t067 represented
the 82.4% (28/34), and the 67.6% (69/102) of the MRSA isolates from
river water and urban effluents, in that order (Table 1). ST125-t067 has
been geographically highlighted in spain representing the major MRSA
genotype associated with nosocomial infections [1,24]. Other MRSA
genotypes such as ST22-t032 and ST5-t002 (Table 1) have also been
associated with human infections [4,22,24]. LA-MRSA were found in
river water and in urban effluents, although typical genotypes such as ST398/
spa t011 or its single locus variant ST1094 were only sporadically found in
our study (Table 1).These results are likely due to the limited impact of
animals in the areas of sampling, close to urban nucleus.
Our data demonstrated that the predominant MRSA and MSSA genetic
lineages detected in urban effluent and river water were human associated
genotypes. This is likely associated with the potential of colonized
individuals to constantly release S. aureus into the environment [5,6,20],
together with the capacity of S. aureus to persist in the water environments
[6,10,25].
Conclusions
Our data emphasize the potential role of anthropogenic activities in
the S. aureus dissemination throughout the water, and highlights the
need to evaluate the circulation and persistence of this pathogen in the
environment and its possible impact for public health.
Acknowledgments
This work was partially supported by the Autonomous Community
of Madrid (S0505⁄AGR-0265; S2009 ⁄AGR-1489) and EU FP7 grant
ANTIGONE (project #278976).
A. Valverde is funded by a postdoctoral fellowship “Juan de la Cierva”
from the Spanish Ministry of Economy and Competitiveness.
Authors wish to thank Dr. Ursula Höfle for help in field sampling and
Dr. J. A. Carriço (Molecular Microbiology and Infection Unit, Instituto
de Medicina Molecular Instituto de Microbiologia, Faculda de Medicina
de Lisboa, Universidade de Lisboa), for helping us using the Web tool
designed to calculate Simpson’s Index of Diversity (Simpson’s ID) and
confidence intervals, and the technicians María García, Estefania Rivero
and Carolina Castilla (VISAVET) for their excellent technical assistance.
No funding for covering the costs to publish in open access was
received.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors
had no role in the design of the study; in the collection, analyses, or
interpretation of data; in the writing of the manuscript, and in the decision
to publish the results.
Article Information
Aritcle Type: Research Article
Citation: Porrero MC, Valverde A, Mateos A, Cantón
R, Gortázar C, et al. (2016) Staphylococcus aureus
Genetic Lineages Found in Urban Effluents and River
Water. Int J Water Wastewater Treat 2(2): doi http://
dx.doi.org/10.16966/2381-5299.117
Copyright: © 2016, Porrero MC, et al. This is an
open-access article distributed under the terms
of the Creative Commons Attribution License,
which permits unrestricted use, distribution, and
reproduction in any medium, provided the original
author and source are credited.
Publication history:
Received date: 06 Oct 2015
Accepted date: 25
Jan 2016
Published date: 01 Feb 2016




