
Figure 1: CLART-StripR -CS platform in the form of an 8-well strip.


Muhammet Guzel Kurtoglu1* Oya Akkaya1 Ayse Ruveyda Ugur1 Edibe Nurzen Bozkurt2 Nurettin Kaya3
1Konya Training and Research Hospital, Department of Medical Microbiology, Konya, Turkey*Corresponding author: Muhammet Guzel Kurtoglu, Konya Training and Research Hospital, Department of Medical Microbiology, Konya, Turkey, Tel: 00903323236709; E-mail: kurtoglumg@gmail.com
Viruses are substantial causes of acute respiratory infections (ARI) worldwide with an estimated rate of 20-60%. The aim of this study is to provide prompt identification of the respiratory viral pathogens in pediatric and adult patients with symptoms of respiratory tract infections by microarray-based multiplex PCR assay and to investigate the frequency of the causative agents according to ages and the seasons.
Between January 2013 and December 2015, 1290 patients (aged 1 month-75 years) diagnosed with ARI were included in the study. Human respiratory virus (RSV), human rhinovirus (HRV), influenza virus (IV), human bocavirus (HBoV), parainfluenza virus (PIV), human metapneumovirus (HMPV), adenovirus (ADV), and human coronavirus-229E (HCoV-229E) were investigated in respiratory specimens by using microarray-based reverse transcriptase PCR (M-PCR).
Of 1290 patients, 1110 (86%) were children and 180 (14%) were adults. At least one causative agent was identified in 750 (58%) samples. Of the 750 positive samples, single virus infection rate was 76% and co-infection rate was 24%. Dual viral agents were detected in 165 specimens and triple viral agents were detected in 15 specimens. Total positivity rates of samples for years 2013, 2014 and 2015 were 41%, 33%, and 26%, respectively. Most of the positive specimens were delivered from the Pediatrics Clinic (94%) and 64.4% of the patients with viral ARI were ≤2 years of age. Positivity rates were increased in winter (40%) and spring (39%) seasons. The most frequent viruses detected were as follows; RSV (47%), HRV (15%), IV (10%), HBoC and PIV (8%), HMPV and ADV (%5), EV and HCoV-229E (%1).
Respiratory tract infection, virus, pathogen, microarray, Multiplex PCR
Viruses are substantial causes of acute respiratory infections (ARI) worldwide with an estimated rate of 20-60% of all cases [1-3]. The most common viruses accountable for ARI are; influenza A and B (IAV and IBV), respiratory syncytial virus (RSV), parainfluenza virus (PIV), adenovirus (ADV), and human rhinovirus (HRV). In recent years, emerging viruses such as human metapneumovirus (HMPV) and human coronaviruses (HCoV) have been implicated in ARI [4].
The annual incidence of pneumonia is reported at rates ranging from 0.28% to 1.16% globally. The incidence and mortality rate of ARI, insisting to be among the top three mortal causes, are increased at early in life and advanced age [5]. The World Health Organization (WHO) reported the mortality rate of ARI 13-15% in children less than 5 years of age in middle income countries in 2014, pointing out that its overall mortalitity rate is higher than most infectious diseases such as AIDS, malaria and measles, even than their sum [6].
The current vaccines do not provide effective prevention against respiratory tract infections caused by viruses. Therefore, useful control and treatment measures should be developed against viral respiratory tract infections. Differences in geographical, climate, and socioeconomic factors necessitate different methods to avoid infections caused by different viruses [7-10].
RSV of Paramyxoviridae family is an enveloped virus with a singlestranded RNA genome. RSV strains were divided into two main groups; RSV-A, and RSV-B depending on their antigenic and genetic diversity RSV is an important etiologic agent of bronchiolitis and pneumonia in infants and young children. RSV is also an important cause of morbidity and mortality in adults. HRV and enterovirus (EV), belonging to Picornaviridae, may also cause upper and lower respiratory tract infections in children. They are non-enveloped viruses with a single-stranded RNA genome. There are more than 100 serotypes of HRV. Twelve species and more than 100 subtypes have been identified in enterovirus genus. Influenza viruses (Orthomyxoviridae) are enveloped viruses with a single stranded and segmented RNA genome. Para influenza viruses are members of the Paramyxoviridae family and possess an envelope and a single stranded RNA. They cause upper and lower airway infections such as rhinitis, otitis, laryngotracheobronchitis, and pneumonia. Human bocavirus (HBoV), which causes acute respiratory infections, was identified in 2005 as a new parvovirus of the Parvoviridae family. It is an non-enveloped and single-stranded DNA virus. An enveloped RNA virus, HMPV can cause acute respiratory tract infections in children and in adults. ADV is a non-enveloped double-stranded DNA and can cause respiratory tract infections in addition to gastroenteritis, cystitis and meningoencephalitis. Up to now, over 50 serotypes of ADV have been identified. HCoV is a single stranded RNA virus and implicated in respiratory tract infections, enteritis, cardiovascular infections and neurological defects [11].
RSV, IAV, IBV, ADV, PIVs 1-3, and HMPV can be identified by direct fluorescent antibody and cell culture methods. In addition to these aforementioned viruses, HRV, HCoV, HBoV, PIV 4, and RSV serotype A and B can be detected by molecular methods. Recently, multiplex polymerase chain reaction (M-PCR) methods in which many respiratory viruses are detected at the same time have also been widely used in laboratories for early diagnosis of ARI [12,13].
There are few epidemiological studies investigating the frequency of the viral pathogens causing ARI in different age groups, especially in childhood in Turkey. The aim of this study is to provide prompt identification of the respiratory viral pathogens in pediatric and adult patients with symptoms of respiratory tract infections by microarray-based multiplex PCR assay and to investigate the frequency of the causative agents according to ages and the seasons.
Nasopharyngeal brush samples taken from patients who were consulted to various clinics, mainly to Pediatric Clinics and Clinic for Chest Diseases were delivered to the laboratory in the appropriate viral transport medium (HiViralTM Transport Medium, HIMEDIA, India) for about 36 months throughout 2013-2015. The samples were stored at -20ºC until the RNA/DNA extraction. For each sample, microarraybased reverse transcription polymerase chain reaction (RT-PCR) assay was performed by using CLART® Pneumo Vir (Genomica, Spain) after RNA/DNA was extracted. Most common nineteen respiratory viruses, RSV type A (RSV-A), RSV type B (RSV-B), HRV, influenza A (subtypes H3N2, H1N1, B, C, and H1N1/2009), HBoV, PIV 1, 2, 3, and 4 (subtypes A, B), ADV, HCoV-229E, Enterovirus (Echovirus), HMPV (subtypes A and B) were detected and characterized by means of the amplification of a specific fragment of the viral genome. An Amplification Internal Control was used to ensure the appropriate execution of the process. After the amplification process was accomplished, the amplicons were detected by using low-density microarray platform: CLART (Clinical Arrays Technology) which makes the hybridization and visualization of precipitates of the products very easy by means of its platform in the form of an 8-well strip (Figure 1) [14].
The rates and ages of male and female patients were calculated. The distributions of the causative viral pathogens were measured according to the ages of patients, the seasons, and the clinics. The frequencies of viruses according to the seasons were estimated by using Z test.

Figure 1: CLART-StripR -CS platform in the form of an 8-well strip.
Patients (aged 1 month -75 years) with fever, sore throat, cough, and difficulty in breathing consulted various Clinics of Konya Education and Research Hospital were recruited between January 2013 and December 2015. After physical examination, 1290 patients were diagnosed with ARI due to the presence of signs and symptoms of ARI, abnormal auscultation and abnormal chest X-ray findings. The most common symptoms were fever (≥38o C), cough, and sore throat. Respiratory specimens were taken from 1290 patients. Of these patients, 684 (53.1%) were male and 606 (46.9%) were female. Majority of the patients were children (86% versus 14%). The mean age was 5.66 (SD=12.81116). The range of age groups were shown in (Table 1).
| Age | ≤2 | 3-5 | 6-18 | 19-59 | ≥60 | Total |
| n (%) | 328 (25.4) | 475 (36.8) | 307 (23.8) | 79 (6.2) | 101 (7.8) | 1290 (100) |
Table 1: The distribution of ages of patients diagnosed with ARI [n (%)]
Respiratory specimens were investigated by multiplex RT-PCR (Figures 2-4). While no viral agent was detected in 540 (42%) of all samples, at least one causative agent was identified in 750 (58%). Of the 750 patients whose samples were positive, 686 were children and 64 were adult patients (Table 2). Viral agents were more frequently detected in winter (40%) and spring (39%). A single viral pathogen was identified in 570 (76%) of all positive specimens while multiple viral agents were co-detected in 180 (24%) samples (Tables 3 and 4). Most of the positive specimens (96%) were delivered from the Pediatric Clinic. Dual pathogens were detected in 165 samples and triple in 15. Among the 19 types of the viruses, RSV was the most frequently detected agent (47%), followed by HRV (15%), and influenza A-B (10%). The rate of multiple viral agents was 24% (Table 4). RSV, followed by influenza virus and adenovirus were the most common viruses in co-infections.
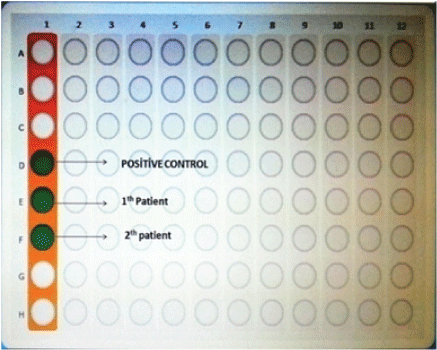
Figure 2: Microarray wells.

Figure 3: Negative results of patients.
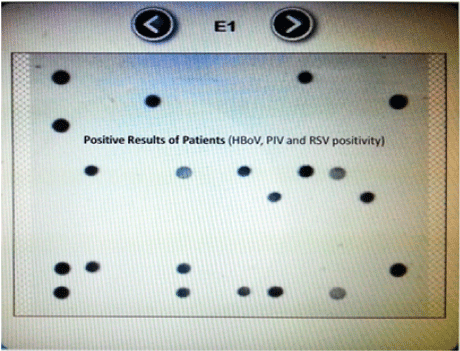
Figure 4: Positive results of patients.
| Age | ≤2 | 3-5 | 6-18 | 19-59 | ≥60 | Total n |
| RSV | 150 (31%) | 90 (53%) | 15 (44%) | 8 (24) | 5 (17%) | 268 |
| HRV | 53 (11%) | 17 (10%) | 5 (15%) | 8 (24) | 3 (10%) | 86 |
| IV | 24 (5%) | 14 (8%) | 8 (23%) | 5 (14) | 6 (20%) | 57 |
| HBoV | 28 (6%) | 8 (5%) | - | 8 (24) | 3 (10%) | 47 |
| PIV | 38 (7.8%) | 5 (3%) | - | - | - | 43 |
| HMPV | 25 (5%) | 3 (2%) | 3 (9%) | - | - | 31 |
| ADV | 19 (4%) | 2 (1%) | 3 (9%) | - | 2 (7%) | 26 |
| EV | 5 (1%) | - | - | - | 1 (3%) | 6 |
| HCoV-229E | 1 (0.2%) | - | - | 2 (5) | 3 (10%) | 6 |
| Co-infection | 140 (29%) | 30 (18%) | - | 3 (9) | 7 (23%) | 180 |
| Total n (%) | 483 (64.4%) | 169 (22.6%) | 34 (4.5%) | 34 (4.5%) | 30 (4%) | 750 |
Table 2: The rates of viruses detected in accordance with the age ranges [n (%)]
| Virus | Winter n (n/216=%) |
Spring n (n/227=%) |
Summer n (n/40=%) |
Autumn n (n/87=%) |
Total n |
| RSV | 126 (58) | 130 (58) | 2 (5) | 10 (12) |
268 |
| HRV | 12 (6) | 30 (13) | 8 (20) | 36 (41) |
86 |
| IV | 23 (11) | 30 (13) | 3 (7) | 1 (1) | 57 |
| HBoV | 11 (5) | 9 (4) | 13 (32) | 14 (16) |
47 |
| PIV | 14 (6) | 0 | 7 (18) | 22 (25) |
43 |
| HMPV | 20 (9) | 10 (4) | 1 (3) | 0 | 31 |
| ADV | 6 (3) | 12 (5) | 4 (10) | 4 (5) | 26 |
| EV | 0 | 4 (2) | 2 (5) | 0 | 6 |
HCoV-229E |
4 (2) | 2 (1) | 0 | 0 | 6 |
Single virus infections |
216 (38) | 227 (40) | 40 (7) | 87 (15) |
570 |
Co-infections |
88 (49) | 65 (%6) | 12 (7) | 15 (8) | 180 |
Total n (%) |
304 (40) | 292 (39) | 52 (7) | 102 (14) |
750 |
Table 3: The distribution rates of viruses detected in accordance with the seasons [n (%)]
| Virus | 2013 n (n/235=%) |
2014 n (n/180=%) |
2015 n (n/155=%) |
Total n |
| RSV | 97 (41) | 98 (54) | 73 (47) | 268 |
| HRV | 47 (20) | 20 (11) | 19 (12) | 86 |
| IV | 24 (10) | 15 (8) | 18 (12) | 57 |
| HBoV | 18 (8) | 18 (10) | 11 (7) | 47 |
| PIV | 16 (7) | 12 (7) | 15 (10) | 43 |
| HMPV | 15 (6) | 8 (4) | 8 (5) | 31 |
| ADV | 12 (5) | 6 (3) | 8 (5) | 26 |
| EV | 2 (1) | 1 (1) | 3 (2) | 6 |
| HCoV-229E | 4 (2) | 2 (2) | 0 | 6 |
| Single infection | 235 (41) | 180 (32) | 155 (27) | 570 |
| Co-infection | 70 (39) | 70 (39) | 40 (22) | 180 |
| Total n (%) | 305 (41) | 250 (33) | 195 (26) | 750 (100) |
Table 4: The distribution rates of viruses detected in accordance with the years [n (%)]
Positivity rates were increased in winter (40%) and spring (39%) seasons. Respiratory syncytial virus was detected most frequently in winter and spring seasons, HRV was detected most frequently in the fall, and IV was detected most frequently in the spring season (Table 3). Total positivity rates of samples for years 2013, 2014 and 2015 were 41%, 33%, and 26%, respectively (Table 4). There was a significant difference according to Z-Test statistics (Z value>Z (0.05) table value).
Respiratory tract infections are the leading illnesses among the most common infectious diseases worldwide. Therefore, ARI imposes huge economic burden due to the factors such as treatment costs, hospitalization expenditures, and labour loss arising from the high attack rates, as well as its high morbidity and mortality rates. Also, it still remains as a huge problem as it is one of the leading causes of child loss under 5 years of age. Lower respiratory tract infections (LRTI) are less common than upper respiratory tract infections (URTI). However, the economic impact of LRTI is higher than that of URTI. 1-13% of patients diagnosed with LRTI are hospitalized. Estimated treatment cost of viral LRTI in children is about 2.4 billion dollars annually [15].
The most common viral pathogens we detected in children less than 5 years of age were RSV, HRV, PIV, IV, and HBoV, followed by HMPV, ADV, EV, and HCoV. The most prevalent viruses in adults (≥19 years) were RSV, HRV, IV, and HBoV, followed by HCoV, ADV, and EV. It is noteworthy to mention that PIV and HMPV were not detected in adults.
Consistent with our study, Akkaya et al. [16] reported that HRV was the second most frequent cause of ARI in children, but RSV and IV were the two most common viruses in adults. The extensive influenza vaccination programs may be the cause of relative rise in RSV related ARIs [16,17].
Hak et al. [18] reported the most frequent viral pathogens causing respiratory infections in Holland as follows: IV, PIV, RSV, and ADV. It has also been reported in different studies that PIV is the prominent viral pathogen, especially by causing re-infections in young children with incomplete immunity [19,20]. The most frequent causative viral agents were reported as IV (30%), PIV (18%), RSV (16%), ADV (15%), HRV (11%), and HMPV (1.5%) in China [21].
In our study, the detection rates of HRV and HMPV were increased slightly in 2013, the detection rates of RSV and HBoV in 2014, and the detection rates of IV and PIV in 2015 during the time period of 2013- 2015. Consistent with the literature, we found that RSV and IV were more frequent in winter and in the spring seasons than in autumn and summer. In contrast, HRV was most common during the fall and summer seasons and its frequency was declined during the winter and spring. While detection rates of HBoV, ADV, and enterovirus were increased during the summer, PIV rates were greatest in autumn and not detected during spring. Furthermore, although their overall frequencies were very low throughout 2013-2015, enterovirus was not detected in autumn and winter, and HCoV was not detected in summer and autumn.
Consistent with our study, different investigators have reported that respiratory viruses were more frequent in winter months in the northern hemisphere, and that RSV and IV were most common viral pathogens detected [16,21-26]. The detection rates of HCoV, HMPV, and HBoV were reported in various studies to be as low as 10-15%, 2-12%, and 1-5%, respectively [3,18,23,24,27]. Interestingly, all of the 31 HMPV were detected only in children in this study. HMPV was detected in children in other studies, too [21]. In our study, ADV was more frequent in children than in adults, consistent with various studies in the literature [9,28,29]. ADV was found to be endemic in all age groups throughout the year, especially in immunosuppresive patients [30-32].
HCoV is an important respiratory pathogen in adults [33]. We detected HCoV only in a pediatric patient, who was under 2 years of age, and in five adult cases. There are many studies reporting that HCoV is more common in adults than in children [21,34,35]. There are also studies reporting that HCoV leads to ARI more commonly in immunosuppressive patients [32,36,37].
In this study, the single virus infection rate was found to be 76%, which has been frequently observed in the pediatric age group (77%). Many investigators have reported that single infections are more often in the pediatric age group, with rates ranging from 35.15% to 36.6% [18,21,38-44].
It has also been shown that multiple viruses can co-infect the respiratory tract by using M-PCR technology developed recently with high specificity and sensitivity [45-50]. The co-infection rate in our study was 24%. RSV was the most frequent component of the co-infections, followed by IV, and ADV. In different studies, co-infection rates have been varied at a rate of 5-62% and it has been reported that RSV is the most frequent component of these infections, followed by ADV, HBoV, and IV [18,22,51,52]. These results, which have been found by various researchers, were consistent with our study, and co-infections have been reported mostly in the childhood age groups [22,24].
One of the endpoints that biotechnology conceptually arrives is the microarray method. It is based on the principle allowing the visualization of the targeted genome on a simple chip. It also allows to determine interactions of many genes. Microarray technology was originally derived from the “Southern Blotting” technique. This fast and automated technology is also used in gene expression analysis, genetic and mutation analysis, environmental research, antimicrobial resistance gene analysis, and in identifying infectious agents as well [53,54]. In two studies, out of 511 samples in which viral pathogens are investigated, 349 were identified as positive by using microarray, consistent with the results obtained by PCR and DNA sequencing methods [55]. Because the microarray method is an automated system, it has minimized man-made errors [56]. Despite of many advantages, it must be known that there are some disadvantages of microarray method, such as performing too much data at once, taking time to analyze all results, complicated and interpretive results that are usually not quantitative enough, and still being an expensive technology [57].
In conclusion, in this study, we detected viral pathogens in patients with the initial diagnosis of ARI at a rate of 58% prevalence, most of whom were pediatric age group (86%). Viral respiratory tract infections were found to be more frequent in winter and spring seasons. Most of the co-infections were seen in the pediatric patients. RSV, HRV, IV, and HBoV were found to be the most frequent pathogens overall. Prompt and simultaneous identification of all the causative agents is crucial for the prognosis of the disease, as well as for the declining the treatment costs and hospitalization time. Microarray-based multiplex PCR method, which has a high sensitivity and specificity, allows detection of multiple pathogens simultaneously and early diagnosis, prevents unnecessary antibiotic use, and decreases morbidity and mortality rates associated with ARI. Therefore, M-PCR can be considered as an alternative method to conventional diagnostic assays. Early diagnosis of viral infections is important because it reduces the cost of treatment and the length of hospitalization. We attributed the reason of slight differences in the frequencies of viruses to differences in demographic characterization of patient groups, seasonal variations, and to specific epidemic differences.
A rapid and accurate diagnosis of viral ARI is important, especially in hospitalized patients, to avoid unnecessary antibiotic use, to take necessary infection control measures, and to allow administration of a specific treatment when necessary if a specific antiviral therapy exists for a causative agent. In addition, with the aid of identification of viral pathogens, improvements in information allow prediction of causative agents in accordance with the factors such as the age of the patient and the season. Therefore, we recommend the use of M-PCR in the diagnosis of hospitalized pediatric patients with ARI.
Download Provisional PDF Here
Article Type: Research Article
Citation: Kurtoglu MG, Akkaya O, Ugur AR, Bozkurt EN, Kaya N (2017) Microarray-Based Molecular Detection of Viral Pathogens associated with Respiratory Infections. J Infect Pulm Dis 3(1): doi http://dx.doi.org/10.16966/2470-3176.122
Copyright: © 2017 Kurtoglu MG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access