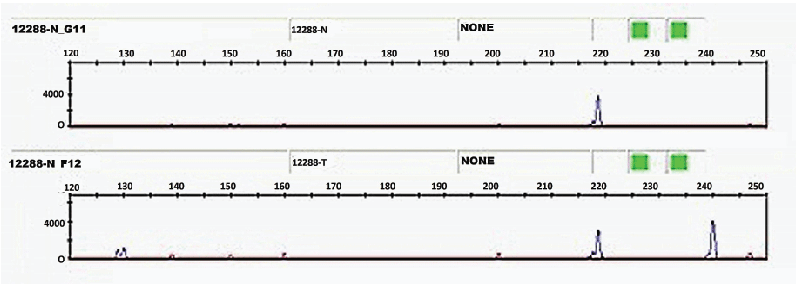
Figure 1: Electrophoregrams showing normal DNA genotyped O/O (top) and tumor DNA genotyped A/O (below) of one case (indicate sample ID).
S Zouine1,2,* N Zaid3 A Bakhchan4 Y Zaid5 K Kojok6 N Tahiri Jouti7 F Marnissi8 N Habti1,2
1Laboratory of Biotechnology and Experimental Medicine- Faculty of Medicine and Pharmacy Casablanca - Hassan II University of Casablanca, Morocco*Corresponding author: Soukaina Zouine, Laboratory of Biotechnology and Experimental Medicine, Laboratory of Hematology, Cellular and Genetic engineering - Faculty of Medicine and Pharmacy Casablanca - Hassan II University of Casablanca, Morocco, E-mail: soukaina.zouine@gmail.com
Introduction: Expression of incompatible A antigen in malignant cells of subjects with blood type O has been reported in several studies. While this phenomenon has been observed in 10-20% of blood group O patients, the genetic mechanism responsible for incompatible A antigen expression remains unknown.
Material and Methods: Genomic DNA extracted from 22 mammary waxed blocks corresponding to 7 patients who have already shown an expression of incompatible A in tumor served as a template for ABO genotyping performed by fluorescent, duplex SSP-PCR. Exons 6 and 7 of ABO blood group gene were also sequenced by next-generation sequencing (NGS).
Results: 4/7 patients were genotyped O/O in normal and tumor DNA, 3/7 cases were genotyped O/O in normal DNA and A/O in tumor DNA. New frameshift mutation 320delA was found creating stop codon in exon 6 of an allele O. In 3/7 cases of tumor tissue expressing the incompatible A antigen, deletion 320 is removed, the stop codon resulting no longer exists, the reading frame is not altered and therefore a glycosyltransferase A is synthesized.
Conclusion: Incompatible A antigen expressed in patients with phenotype O is derived from an allele O deletional at position 320 by a functional and immune reactive protein similar to the A transferase in exons 6 and 7. Despite its biological function poorly known in the biogenesis of cancer, the expression of A antigen on the surface of tumor cells could make the antigen clinically useful as a biomarker of tumor prognosis.
ABH antigens; Breast cancer; Incompatible A antigen; non deletional O; 320delA
The synthesis of ABH antigens requires expression of transferase enzymes encoded by the ABO gene [1,2]. This gene is located on the distal long arm of human chromosome 9. It is composed of seven exons and encodes a 354-amino acid glycosyltransferase. A, B and O types and subtypes result from genetic variants within the gene, which affect either the expression or activity of the enzyme leading to variations in the quality and/or amount of blood group antigens detectable at the surface of red blood cells (RBCs) [3-6].
Genetic modification has been reported to be common in several cancers derived from epithelial cells [7,8]. Breast cancer often triggers changes in the glycosylation status occurring in the membrane glycoconjugates, such as BRCA1, BRCA2, and the Thomsen-Friedenreich sialyl-Lewis antigen, which are involved in cancer development. Among glycoconjugates, histo-blood group ABH antigens are also assumed to be implicated in various cancers [9-12].
Several studies have demonstrated that cells normally expressing ABH antigens may partially lose ABH expression when a malignant process occurs. Expression of A and B antigens has been established as a favorable prognostic factor, as their reduction or complete deletion is correlated with poor prognosis [13]. De novo ABH antigen expression identical to embryonic life is also observed in some solid tumors, including colorectal cancer [6,14-16]. Some subjects with RBC blood group O phenotype may present an incompatible A antigen expressed in their malignant cells [17].
There are few studies exploring the mechanism of synthesis of an incompatible A antigen. Clausen and colleagues suggested that point mutations may occur in the O allele, restoring the reading frame and resulting in the production of an A-like transferase [18,19].
However, the question arose as to how a given mutation may be a common feature of different types of cancer in different individuals. Other hypotheses have been proposed, such as the use of a second start codon in the O allele, which may result in the production of a partially, N-truncated enzymatically active protein or the activation of a pseudo-gene. In addition, alternative splicing would occur and remove exon 6 carrying the c.261G deletion resulting in the production of a transcript missing 135 base pairs and a truncated protein lacking 33 amino acids [20].
With the discovery of the O allele devoid of the common deletion (O03) and at a frequency of 7.3 to 8%, it is assumed that this allele contributes to the expression of incompatible A antigen.
We have screened 7 cases suffering from breast cancer with RBC phenotype O, who were found to express blood group A antigen in tumor detected by immunohistochemistry in order to explore the mechanism involved in A incompatible expression.
The study was conducted according to the Declaration of Helsinki, evaluated and approved by the Ethic Committee for Biomedical Research of Rabat (CERBR) of the Faculty of Sciences, Rabat-University Mohammed V.
Seven RBC phenotype O cases suffering from breast cancer, who showed an expression of incompatible A antigen in the tumor by immunohistochemistry, were selected for the study. Samples, i.e. patientmatched pairs of normal (N)/tumor (T) tissues, were obtained from mammary tumorectomy or mastectomy from the Pathology Department, University Hospital Ibn Rochd (Casablanca, Morocco). All samples were fixed in formalin, embedded in paraffin and contained representative portions of both tumor and adjacent normal mammary tissue, all expressing A and H antigens, respectively.
For each sample, the tumor and adjacent noncancerous tissues were circled, scraped with a scalpel and placed in microtubes for dewaxing with xylene and absolute ethanol.
Extraction of genomic DNA from fixed mammary tissue was carried out by using the manual QIAamp DNA FFPE Tissue MinElute® kit (Qiagen), as recommended in the “tissue” protocol provided by the manufacturer. Extracted DNA was eluted in 60 μL TE Buffer and stored at -20°C before use.
Nucleotide positions c.261 (PCR 1) and c.703 (PCR 2) of the ABO gene were analyzed by fluorescent, duplex sequence-specific primer (ASP)- PCR (Table 1). PCR reactions were performed in a final reaction volume of 10 µL with 1X HotStarTaq PCR Master Mix (Qiagen), 0.4 µM of each primer, 0.5 µM of the universal U-FAM primer (Table 1) and 2 µL of purified genomic DNA.
PCR |
Theoric size (pb) |
Allele |
Primers |
|
Sequence (5’→3’) |
||||
1 |
243 |
AorB |
ABO_261G_Fa |
GTTTCTTCAGCCAAAGGTCTGACACCCCGGAAGGATGTCCTCGTGGTGA |
|
222 |
O |
ABO_261X_Fa |
GTTTCTTGTGGAAGGATGTCCTCGTGGTAC |
|
- |
- |
ABO_261C_RuFb |
GTCGTAGTCGACGACCGTTAAATGTCCACAGTCACTCGCCACT |
2 |
153 |
B |
ABO_703A_Fa |
GTTTCTTGTGGAGATCCTGACTCCGCTCCTCGGCACCCTGCACCACA |
|
133 |
A or O |
ABO_703G_Fa |
GTTTCTTGTTCGGCACCCTGCACCCGG |
|
- |
- |
ABO_703X_RuFb |
GTCGTAGTCGACGACCGTTAGAAATCGCCCTCGTCCTTGG |
- |
- |
- |
U-FAM |
6-FAM-GTCGTAGTCGACGACCGTTA |
Table 1: Primers for fluorescent duplex PCR-SSP
aSpecific forward primers with genotyping sequence GTTTCTT (underlined).
bCommon reverse primers containing the single sequence GTCGTAGTCGACGACCGTTA (underlined).
Amplified PCR products (0.5 µL) were mixed with GeneScan™ 500 ROX™ (Applied Biosystems) in deionized formamide Hi-Di™ (Applied Biosystems). Products were denatured at 95°C for 5 min and then placed on ice for 5 min. The single-stranded DNA fragments were then separated by capillary electrophoresis in a 3130xl genetic Analyzer (Applied Biosystems) and the resulting electropherograms were analyzed with GeneMapper® Software. Genotypes are finally identified by interpreting PCR 1 and 2 amplification results.
Exons 6 and 7 of the ABO gene were sequenced by using genomic DNA extracted from both normal and tumoral cells
We obtained a minimal depth of the cover of 9X, which provided sufficient depth to analyze SNPs. Depth blankets >5X are significant. Sequences obtained were aligned with the reference genome and analyzed using the NCBI databases and ENSEMBL to confirm the effect of new mutations on the protein. We have sought in the database if the mutations found were already described. We also conducted a study of the literature for each of the mutations.
Here, we report only mutations that are different between normal and tumor DNA. Mutations were confirmed by PCR-based Sanger sequencing.
Among the 7 cases, 4/7 were genotyped O/O in both normal and tumor DNAs, while 3/7 were genotyped O/O in normal DNA and A/O in tumor DNA (Table 2).
Sample ID |
Tissue |
Tissue phenotype (IHC) |
Genotypea |
Nucleotide positionsb |
||
c.261 |
c.320 |
c.740 |
||||
1, 2, 3, 4 |
Normal |
H |
O/O |
G/delG |
A/delA |
A/G |
|
Tumor |
A |
O/O |
G/delG |
A/delA |
A/G |
5, 6, 7 |
Normal |
H |
O/O |
G/delG |
A/delA |
A/G |
|
Tumor |
A |
A/O |
G/delG |
A/A |
A/A |
Table 2: Genotype-phenotype in normal and tumor tissues in seven incompatible A antigen-expressing patients with blood type O.
aGenotype determined by fluorescent, duplex SSP-PCR.
bNucleotide positions determined by sequencing.
Changes are underlined; del: deletion.
ABO gene sequencing of seven patients expressing an incompatible A antigen in the tumor revealed the presence of two new different mutations, which have been reported neither in database nor in the literature, to our knowledge. The first mutation is a single base deletion in exon 6, i.e. c.320delA, which potentially results in the production of a nonfunctional product lacking the catalytic active site (p.Glu107GlyfsX12).
The second mutation is a single-base substitution in exon 7, i.e. c.740A>G, which is supposed to replace a glutamate residue by a glycine residue (p.Glu247Gly).
These two SNPs were present in the normal DNA of the seven patients (i.e. cases 1/N to 7/N), as well as in tumor DNA of 4/7 patients (i.e. cases 1/T to 4/T). Conversely, in 3/7 patients (i.e. cases 5/T to 7/T) nucleotide A at position 320 was not deleted and mutation c.740A>G was absent in tumor DNA (Table 3).
In addition, the common deletion of allele O, 261delG was present in the DNA of all patients.
Case number |
ABO Genotype |
c.320delA |
c.740A>G |
1/N |
O/O |
+ |
+ |
1/T |
O/O |
+ |
+ |
2/N |
O/O |
+ |
+ |
2/T |
O/O |
+ |
+ |
3/N |
O/O |
+ |
+ |
3/T |
O/O |
+ |
+ |
4/N |
O/O |
+ |
+ |
4/T |
O/O |
+ |
+ |
5/N |
O/O |
+ |
+ |
5/T |
A/O |
- |
- |
6/N |
O/O |
+ |
+ |
6/T |
A/O |
- |
- |
7/N |
O/O |
+ |
+ |
7/T |
A/O |
- |
- |
Table 3: Mutations in exons 6 and 7 among seven patients.
+: presence of mutation;
-: absence of mutation.
Incompatible A antigen was present and located only in the tumor while the adjacent breast tissue that appears normal was not labeled with the anti-A and shows no enzymatic activity, confirming that the incompatible antigen expression is a phenomenon linked to cancer rather than the individual [21-24].
4/7 patients were genotyped O/O in normal and tumor, 3/7 patients genotyped O/O in the normal and A/O in the tumor. This result is not in agreement with David L’s study which has shown that all cases expressing A antigen are genotyped O/O with common deletion 261delG [25].
In order to investigate possible mutations responsible for A antigen expression and to answer the hypothesis supporting that A antigen is from a non deletional O allele, we sequenced the last two exons 6 and 7 of the ABO gene. The sequencing of the normal and tumor DNA of the 7 patients showed the presence of the commune deletion 261delG and two new mutations at position 320 (c.320 delA) in exon 6 and 740 (C.740 A>G).
Our result suggests that normal DNA genotyped O/O is heterozygous in 261, 320 and 740. One allele is deletional at 261 and the second is deletional at 320. The stop codon resulting from deletion 320 would produce a truncated protein incapable of giving a glycosyltransferase activity A or B and thus normal tissue expresses only O antigen. However, in the tumor tissue expressing the A antigen, tumor DNA is heterozygous in 261 while deletion 320 is removed, the stop codon resulting no longer exists and the reading frame is not altered and therefore a glycosyltransferase A is synthesized (Figure 1).

Figure 1: Electrophoregrams showing normal DNA genotyped O/O (top) and tumor DNA genotyped A/O (below) of one case (indicate sample ID).
In contrast, tumor DNA of 4/7 patients genotyped O/O has the same deletions c.320 delA and c.740 A>G while tumor tissue expressed the corresponding incompatible A antigen. By analyzing the expression of H antigen in the normal and tumor tissue, we have observed in one patient (1/4) that expression of H antigen is retained in the tumor tissue which was strongly marked, 80% of cells expressed H antigen versus 40% of cells expressed incompatible A antigen. In this case, sequencing result is probably dependent on the proportion or amount of DNA present in the tissue sampled. In the remaining patients (3/4), H antigen was not expressed on the tumor tissue. Another molecular mechanism could be responsible of incompatible A antigen.
On the other hand, Maaf-Hosseini et al. [26] indicated that non deletional O alleles appear regularly and represent a serious risk for erroneous interpretation in serology and genotyping although they are uncommon. The same team has studied one case of blood donor with the phenotype A low agglutination and genotyped A1/O1. An O allele lacking the common mutation c.261delG and presenting mutation 322C>T, that creates an immediate stop codon after the amino acid 107, has been genotyped as an A1 allele. This allele is assumed to be completely unable to produce an inactive enzyme, but that is not the case. Indeed, the presence of a stop codon does not necessarily mean that the enzyme is inactive (Figure 2).
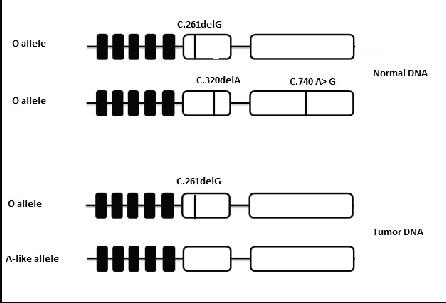
Figure 2: Schematic model of mutations present in O allele and A-like allele.
Two hypotheses have been suggested. There is probably another independent gene capable of initiating biosynthesis transferase as a gene conversion. Moreover, a recombination event could occur in clones of somatic cells in the marrow or outside thereof, and cause double populations within the same person [24,27].
Otherwise, we did not observe significant differences in clinical and pathological features between cases expressing the antigen A-like and other cases. Authors consider that incompatible A antigen expression would be the rejection of cancerous cells due to the natural occurrence of antibodies against blood group antigens of non self [28].
The classic example is that of a small p blood group patient suffering from gastric cancer whose tumor tissue expressed incompatible P antigens. Levine has suggested that the patient was completely restored due to incompatible transfusion P which stimulated the synthesis of anti-P, P2 and pK and therefore the patient’s immunoresponse against incompatible P antigen [29].
It was also reported that women with blood group O having given birth to blood group O children have a relatively high incidence of colorectal cancer compared to women with blood group O having given birth to incompatible children A blood [30]. The low incidence of adenocarcinoma patients in type O individuals could provide an evidence of the importance of incompatible antigen A expression as a trigger for immunologic surveillance against cancer cells.
Our findings suggest the same assumption since antigen A was present in the cytoplasm in all cases. It is recognized that the majority of tumor antigens inducing an immune response in cancer are cytosolic proteins synthesized endogenously and degraded in the cytosol in small peptides associated with the Major Histocompatibility Complex class 1 [31, 32].
Our results allow to conclude that incompatible A antigen expressed in patients with phenotype O is derived in 42.86% cases from an allele O deletional at position 320 by a functional and immunoreactive protein similar to the A transferase in exons 6 and 7. Despite its poorly known biological function in the biogenesis of cancer, the expression of A antigen on the surface of tumor cells could make the antigen clinically useful as a biomarker of tumor prognosis. If tumors expressing the A antigen are recognized and could be suppressed by the immune system, then it could represent an interesting potential target for immunotherapy.
The authors declare having no conflict of interests.
All authors have read and approved the final version of the manuscript.
We are extremely grateful to Pr S. ZAMIATI who has facilitated for us access to Pathology Department.
Download Provisional PDF Here
Article Type: Research Article
Citation: Zouine S, Bakhchan A, Zaid N, Zaid Y, Kojok K, et al. (2016) The non deletional O is involved in incompatible A expression in breast cancer. J Blood Disord Med 2(1): doi http://dx.doi. org/10.16966/2471-5026.113
Copyright: © 2016 Zouine S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access